The BioGateway Resource
A Semantic Systems Biology Database
BioGateway consists of a graph-based database built on Semantic Web principles, a SPARQL endpoint allowing users to query it, and a Cytoscape app which integrates the query functionality directly into your network building workflow.
What is BioGateway?
BioGateway is an initiative that enables a Semantic Systems Biology approach. It provides an entry point to access a data warehouse where biological data is gathered in the form of triples (using RDF). The systems can be queried using SPARQL. The BioGateway system can also be explored using the SPARQL browser. With this browser, SPARQL results can be visually seen as a network of resources.
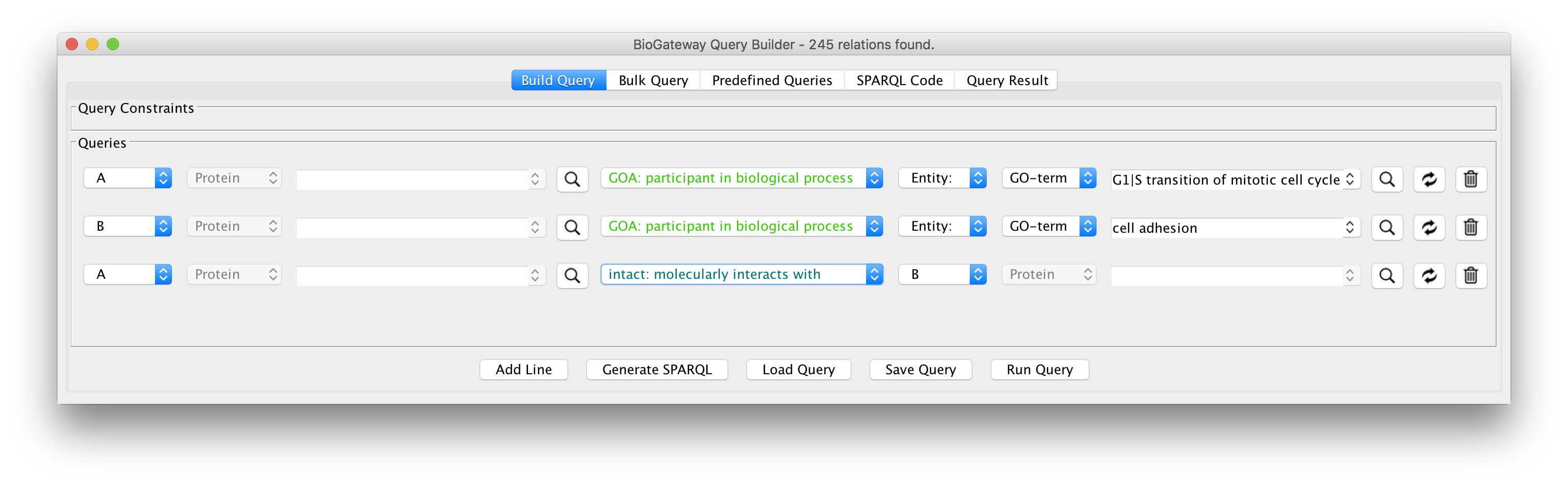
The Cytoscape App
We have developed an app for Cytoscape to allow you to directly integrate the power of our Semantic Knowledge Base into your network building workflow. With the Query Builder tool, you can formulate the topology of what you are looking for, and it will generate the SPARQL query for you.
The query result can then be imported directly into the Cytoscape network you are building – without having to deal with result file formats, incompatible column standards or identifiers.
The BioGateway Database
BioGateway Data model
Overview
The BioGateway triple store provides a unified protein-centric view on biological networks. The data in BioGateway are modeled as directed multi-graphs, not necessarily acyclic, which is a natural choice for representing complex networks.
There are two types of graphs in BioGateway:
A – those that define entities, e.g. proteins, genes, etc.,
B – those that define relations among entities, e.g. protein-protein interaction, protein-disease interactions, etc.
There are three types of nodes in BioGateway:
- Classes: entities in the domain of discourse, e.g. proteins, diseases, etc. (URIs)
- Instances: particular interpretations/views of entities conditioned on the source (URIs, only B type graphs)
- Attributes: qualities, quantities, etc. (literals)
Nodes are connected through multiple types of edges, a.k.a. properties, semantically defined in external ontologies/taxonomies/vocabularies (URIs). Within any given graph a particular property is used within one unique semantic context.
The atomic unit of information (elementary graph) comprises a pair of nodes (subject and object) connected by a directed edge (predicate), commonly known as a triple.
A-type graphs
http://rdf.biogateway.eu/graph/prot
Protein entities (source: ‘http://uniprot.org/uniprot/’, Reference Proteome filtered). This graph forms the core of BioGateway. The entities are identified by their UniParc IDs conditioned on the biological species, chromosome and encoding gene, e.g. ‘http://rdf.biogateway.eu/prot/9606/chr-17/TP53/UPI000002ED67’, thus the corresponding classes are homogeneous with respect to the amino acid sequences. Together with protein classes there are collections of all translation products encoded by a particular gene (essentially sets, but modelled as rdf:Bag due to RDF limitations), e.g. ‘http://rdf.biogateway.eu/prot/9606/chr-17/TP53/’.
http://rdf.biogateway.eu/graph/gene
Gene entities (source: ‘http://uniprot.org/uniprot/’, Reference Proteome filtered). Semantically these entities are defined by the sets of translation products they encode and logistically by the preferred gene names (as used in ‘http://uniprot.org/uniprot/’) conditioned on the biological species and chromosome e.g. ‘http://rdf.biogateway.eu/gene/9606/chr-17/TP53/’. The corresponding entities are not guaranteed to be homogeneous with respect to the nucleotide sequences and modeled as collections (rdf:Bag).
http://rdf.biogateway.eu/graph/taxon
Taxonomic entities (source: ‘http://purl.bioontology.org/ontology/NCBITAXON’) identified by external URIs, e.g. ‘http://purl.bioontology.org/ontology/NCBITAXON/9606’.
http://rdf.biogateway.eu/graph/go
Ontology term entities (source: https://bioportal.bioontology.org/ontologies/GO) identified by external URIs, e.g. ‘http://purl.obolibrary.org/obo/GO_0000122’.
http://rdf.biogateway.eu/graph/omim
Disease entities (source: ‘http://purl.bioontology.org/ontology/OMIM’) identified by external URIs, e.g. ‘http://purl.obolibrary.org/OMIM/151623’.
B-type graphs
All entities are modeled as subclasses of rdf:Statement with instances conditioned on the source.
http://rdf.biogateway.eu/graph/prot2onto
Interactions between proteins and biological processes, cellular components, molecular functions (source: ‘http://identifiers.org/goa’).
e.g:
‘http://rdf.biogateway.eu/prot-obo/9606!chr-17!TP53!UPI000002ED67–GO_0000122’ (class)
‘http://rdf.biogateway.eu/prot-obo/9606!chr-17!TP53!UPI000002ED67–GO_0000122#goa’ (instance).
Defining properties:
‘http://purl.obolibrary.org/obo/RO_0002331’ “involved in” (biological process),
‘http://purl.obolibrary.org/obo/BFO_0000050’ “part of” (cellular component),
‘http://purl.obolibrary.org/obo/RO_0002327’ “enables” (molecular function).
http://rdf.biogateway.eu/graph/prot2phen
Protein-phenotype interactions (currently limited to diseases, source: ‘http://uniprot.org/uniprot/’),
e.g:
‘http://rdf.biogateway.eu/prot-omim/9606!chr-17!TP53!UPI000002ED67–151623’ (class),
‘http://rdf.biogateway.eu/prot-omim/9606!chr-17!TP53!UPI000002ED67–151623#uniprot’ (instance)
Defining property: ‘http://purl.obolibrary.org/obo/RO_0002331’ “involved in” (disease).
http://rdf.biogateway.eu/graph/prot2prot
Protein-protein interactions (source: ‘http://identifiers.org/intact/’)
e.g:
‘http://rdf.biogateway.eu/prot-prot/9606!chr-03!BHLHE40!UPI0000126923–9606!chr-17!TP53!UPI000002ED67’ (class),
‘http://rdf.biogateway.eu/prot-prot/9606!chr-03!BHLHE40!UPI0000126923–9606!chr-17!TP53!UPI000002ED67#intact’ (instance).
Defining property: ‘http://purl.obolibrary.org/obo/RO_0002436’ “molecularly interacts with” (protein).
http://rdf.biogateway.eu/graph/tfac2gene
Interactions between transcription factors and target genes
sources:
‘http://www.tfacts.org’,
‘http://www.grnpedia.org/trrust/’,
‘http://www.lbbc.ibb.unesp.br/htri’,
‘http://signor.uniroma2.it’,
‘http://identifiers.org/intact/’,
‘http://identifiers.org/goa’,
‘http://www.extri.org’,
e.g:
‘http://rdf.biogateway.eu/prot-gene/9606!chr-17!TP53!UPI000002ED67–9606!chr-20!AAR2’ (class),
‘http://rdf.biogateway.eu/prot-gene/9606!chr-17!TP53!UPI000002ED67–9606!chr-20!AAR2#tfacts’ (instance).
Defining property: ‘http://purl.obolibrary.org/obo/RO_0002428’ “involved in regulation of” (gene).
External parental classes
http://www.w3.org/1999/02/22-rdf-syntax-ns#Bag ‘unordered collection’
http://www.w3.org/1999/02/22-rdf-syntax-ns#Statement ‘triple’
http://www.w3.org/2000/01/rdf-schema#Class ‘entity type’
http://www.w3.org/2000/01/rdf-schema#Property ‘edge type’
http://semanticscience.org/resource/SIO_010035 ‘gene’
http://semanticscience.org/resource/SIO_010043 ‘protein’
Properties used in A and B graphs
Object properties
http://www.w3.org/2000/01/rdf-schema#subClassOf ‘is subclass of’
http://www.w3.org/2000/01/rdf-schema#subPropertyOf ‘is subproperty of’
http://semanticscience.org/resource/SIO_000253 ‘has source’ # domain: rdf.bigateway.eu/graph/
http://semanticscience.org/resource/SIO_000772 ‘has evidence’ # range: publications
http://schema.org/evidenceOrigin ‘has evidence origin’ # range: source of metadata
Annotation properties
http://www.w3.org/2004/02/skos/core#prefLabel ‘has name’
http://schema.org/evidenceLevel ‘has evidence level’
Properties used in A graphs
Object properties
http://schema.org/memberOf ‘is member of’
http://purl.obolibrary.org/obo/BFO_0000052 ‘inheres in’ # range: biological species
http://www.w3.org/2004/02/skos/core#closeMatch ‘has close match’ # range: external URIs for genes and proteins
Annotation properties
http://www.w3.org/2004/02/skos/core#altLabel ‘has synonym’
http://www.w3.org/2004/02/skos/core#definition ‘has definition’
Properties used in B graphs
Object properties
http://www.w3.org/1999/02/22-rdf-syntax-ns#type ‘is instance of’
http://purl.obolibrary.org/obo/RO_0002331 ‘involved in’ # range: biological process, disease
http://purl.obolibrary.org/obo/BFO_0000050 ‘part of’ # range: cellular component
http://purl.obolibrary.org/obo/RO_0002327 ‘enables’ # range: molecular function
http://purl.obolibrary.org/obo/RO_0002436 ‘molecularly interacts with’ # range: protein
http://purl.obolibrary.org/obo/RO_0002428 ‘involved in regulation of’ # range: gene
http://www.w3.org/2000/01/rdf-schema#isDefinedBy ‘is defined by’ # range: method
Annotation properties
http://www.w3.org/1999/02/22-rdf-syntax-ns#value ‘has value’ # positive/negative
http://www.w3.org/2000/01/rdf-schema#comment ‘has comment’ # amino acid change
