Application paper
Abstract
The BioGateway App is a Cytoscape plugin that provides easy query access to the BioGateway RDF triple store. BioGateway contains functional and interaction information for proteins from several curated resources. In addition, we have added a comprehensive dataset with regulatory relationships of mammalian DNA binding transcription factors and their target genes, compiled both from curated resources and from a text mining effort. Query results are visualised using the inherent flexibility of the Cytoscape framework, and network links can be checked against curated database records or against the original publication, which offers a potentially efficient mechanism to check text mining results and engage users in community curation.
Reproducing the example from the paper
This page is dedicated to manually reproduce the results from the paper. The first step to do so will be to open the Query Builder.
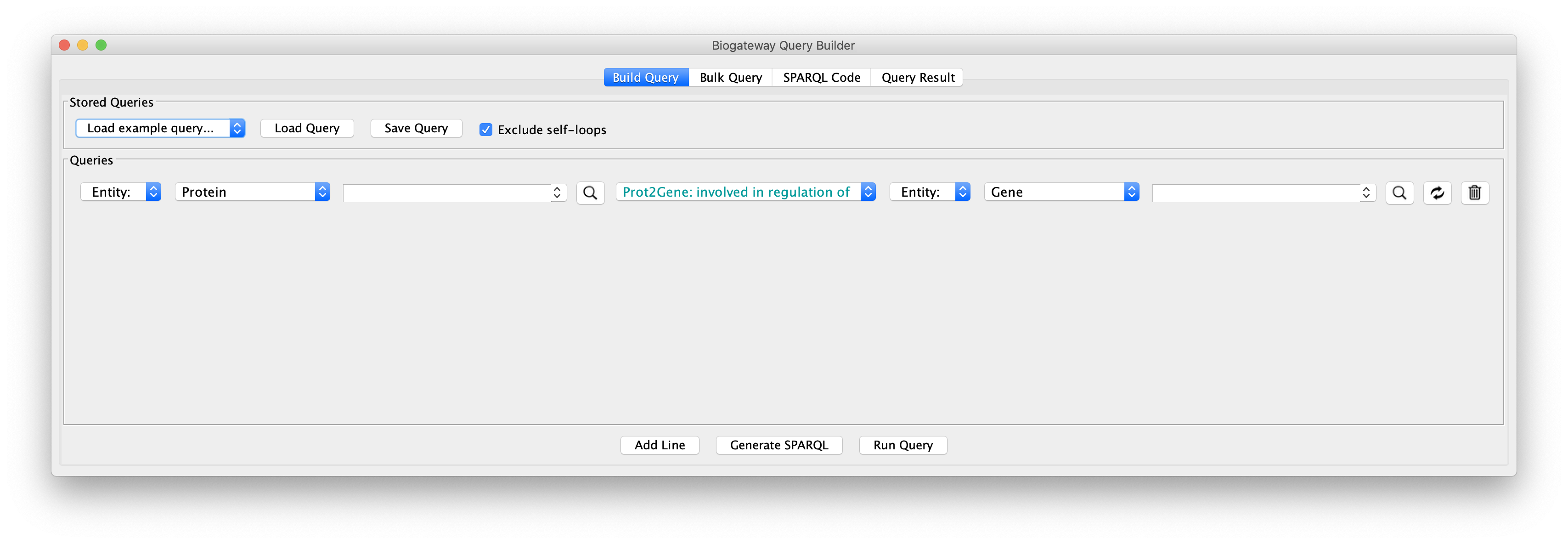
Next, we will create query lines one by one; after completing one the next line can be added by clicking the Add Line button:
- Line 1: set the Subject to Set A, the relation type to GOA: enables molecular function, and the Object to Entity. Start typing protein kinase activity in the text field corresponding to the Object until the autocomplete search engine displays the GO term. Once found, select it by clicking.
- Line 2: set the Subject again to Set A to restrict all proteins involved in Colorectal Cancer to only those with protein kinase activity. The relation type will be UniProt Disease: involved in disease. Finally, the Object will be set to Entity, and the text field will have to contain COLORECTAL CANCER. You will see that there are several hits for Colorectal Cancer. To ensure the reproducibility of this example, select the one written completely in upper case.
- Line 3: set the Subject to Set A, the relation type to Intact: molecularly interacts with, and the Object to Set B. This will find all PPIs containing the proteins found in lines 1 and 2.
- Line 4: set the Subject to Set C, the relation type to UniProt Gene: encodes, and the Object to Set B.
- Line 5: set the Subject to Set D, the relation type to Prot2Gene: involved in regulation of, and the Object to Set C.
- Line 6: set the Subject to Set D, the relation type to Intact: molecularly interacts with, and the Object to Set B.
- Line 7: set the Subject to Set D, the relation type to GOA: involved in biological process. Set the Object to Entity and write angiogenesis in the corresponding text field until the autocomplete engine finds the term. Select it by clicking on it.
The final query should look like the image below:
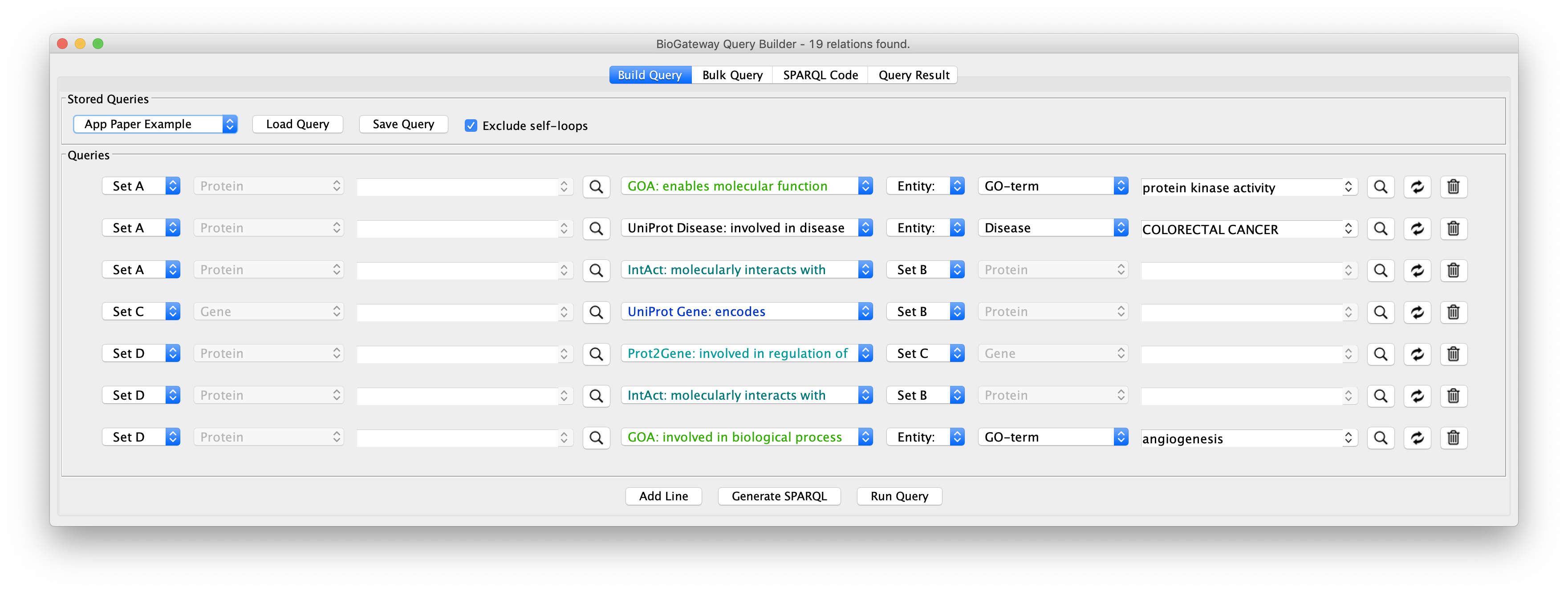
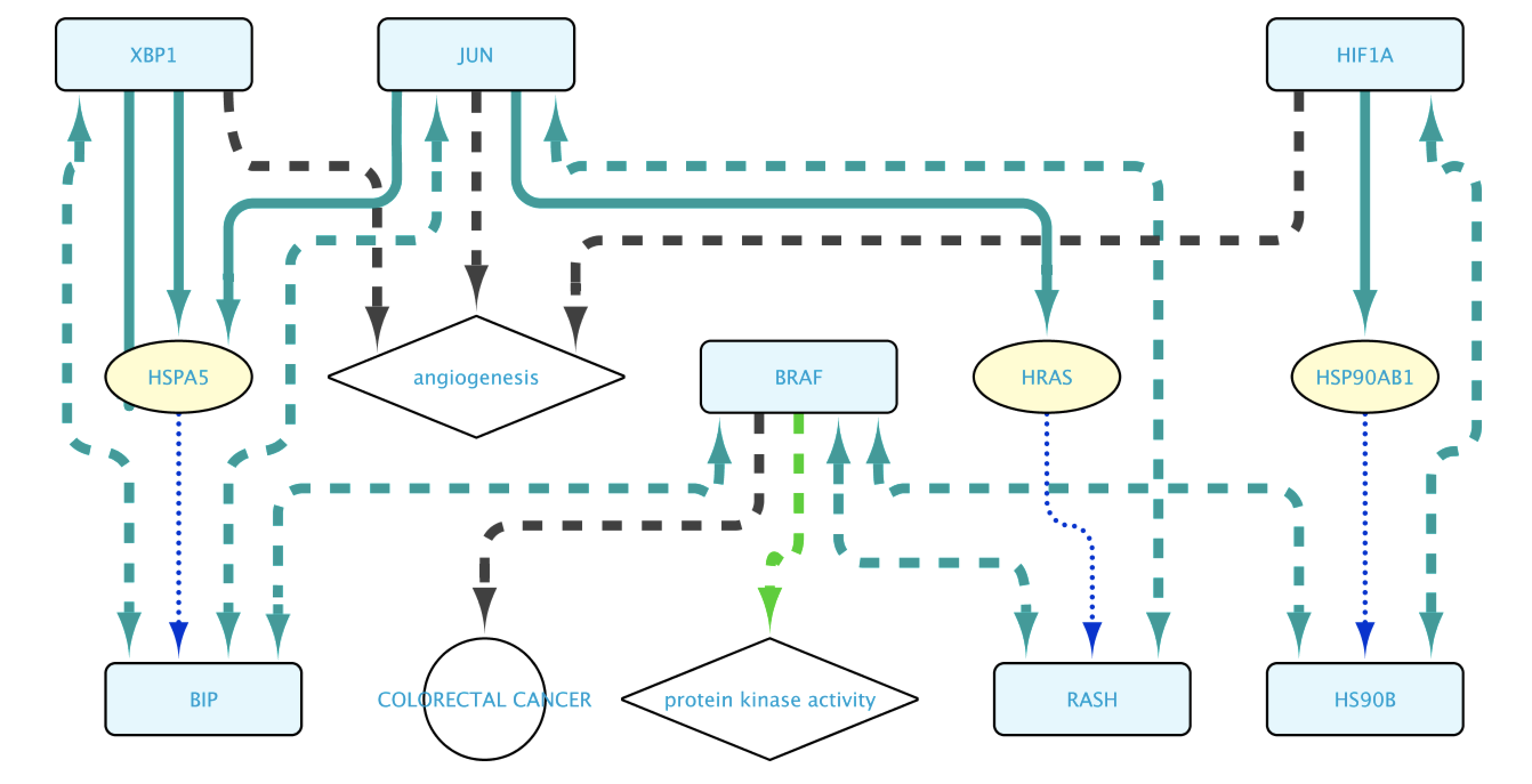
The final steps will be to click the button Run Query. This will open the Query Results tab, where all results have to be selected. Click on the Import to new Network button to generate the Network with the results.
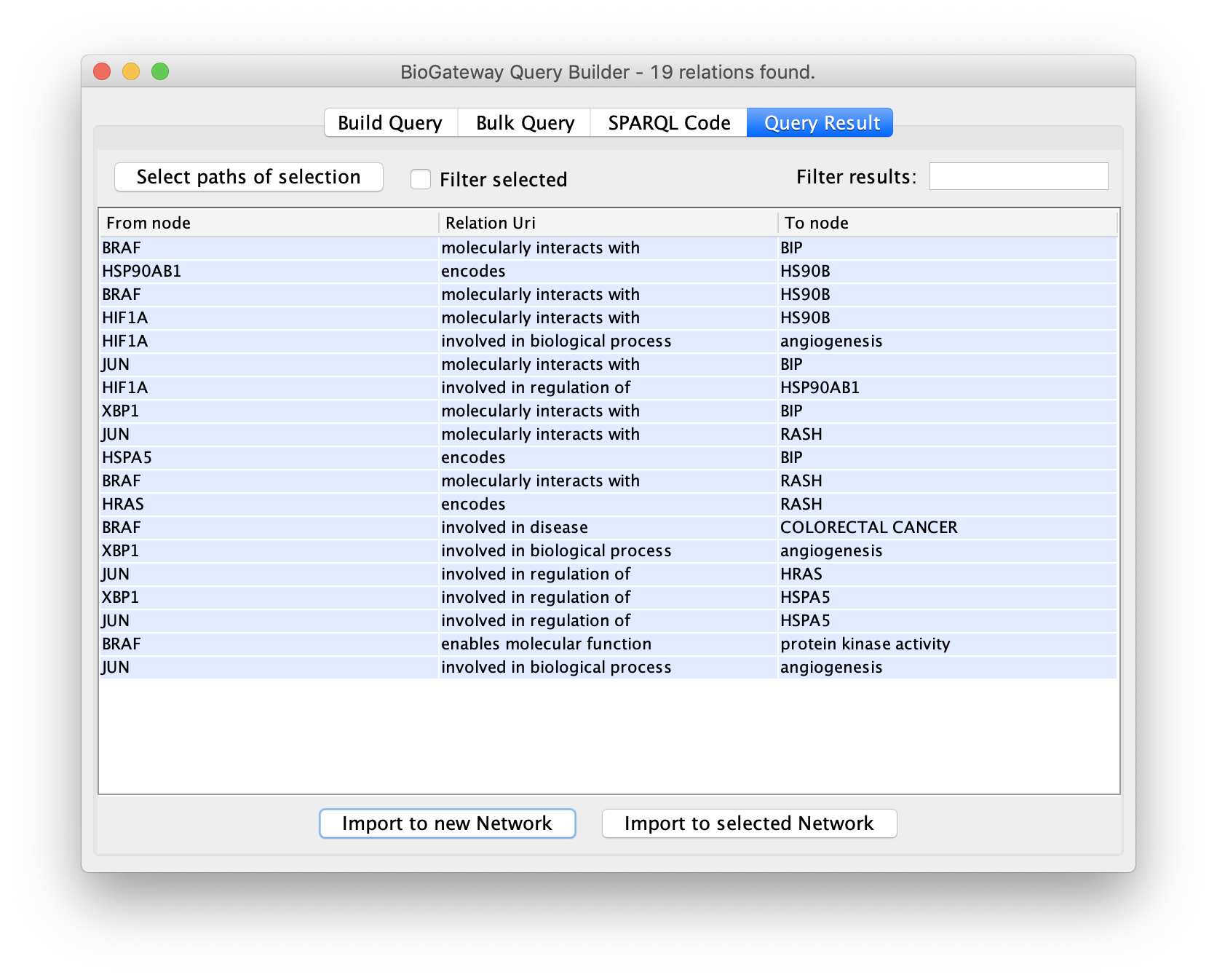
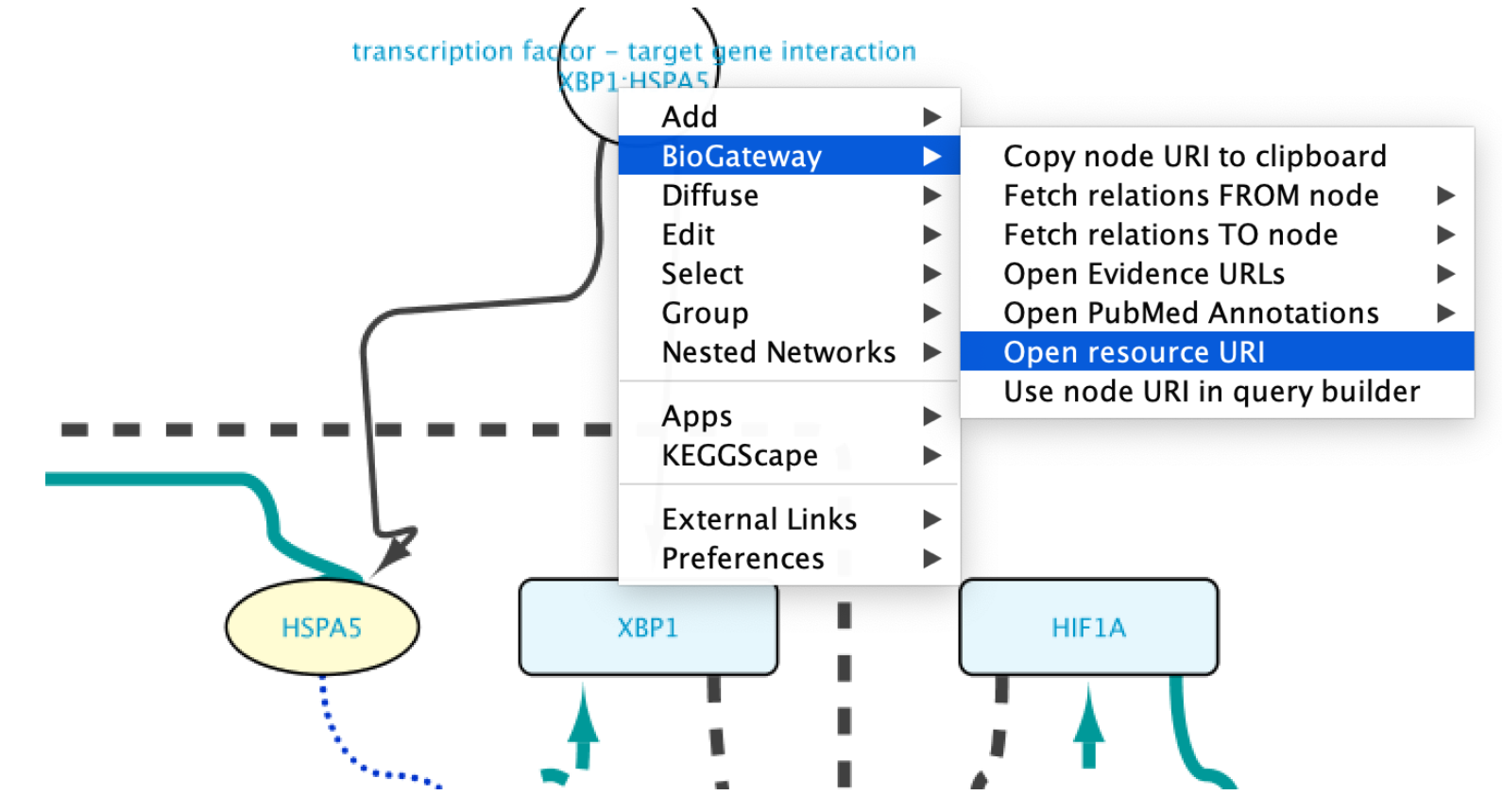
Edges representing relationships can be further explored by double-clicking on them. This allows to check the data supporting them, among other options. In this case, double click on the edge connecting XBP1 and HSPA5. This will create a new node representing the interaction. Right click-on it and select Biogateway > Open resource URI. This will open your web browser, showing the abstracts of the different papers supporting that interaction.