Example use cases
This section describes a diverse set of examples that are intended to illustrate the various functionalities of the Cytoscape Biogateway App and how they can be applied to answer biological questions through a network-based approach. Each of the use cases illustrates an example based on a biological question, translates it into a series of steps to a query and ends with the resulting network. To highlight the added value of the BioGateway App, we also briefly describe an alternative method that should result in (more or less) the same network. A compressed Cytoscape .cys file is provided in the following link to offer the possibility of further exploring the result of each use case. We also provide here a zip file containing all the query files for each of the use cases, so that they can be imported and tweaked for further use.
The App works best in the latest version of Cytoscape (3.7.0 or later). After downloading and installing Cytoscape from www.cytoscape.org you should install the BioGateway App as explained in the Installation page.
Use Case 1 - Exploring Around a Node What is the biological context around the protein GTPase KRas encoded by the KRAS gene?
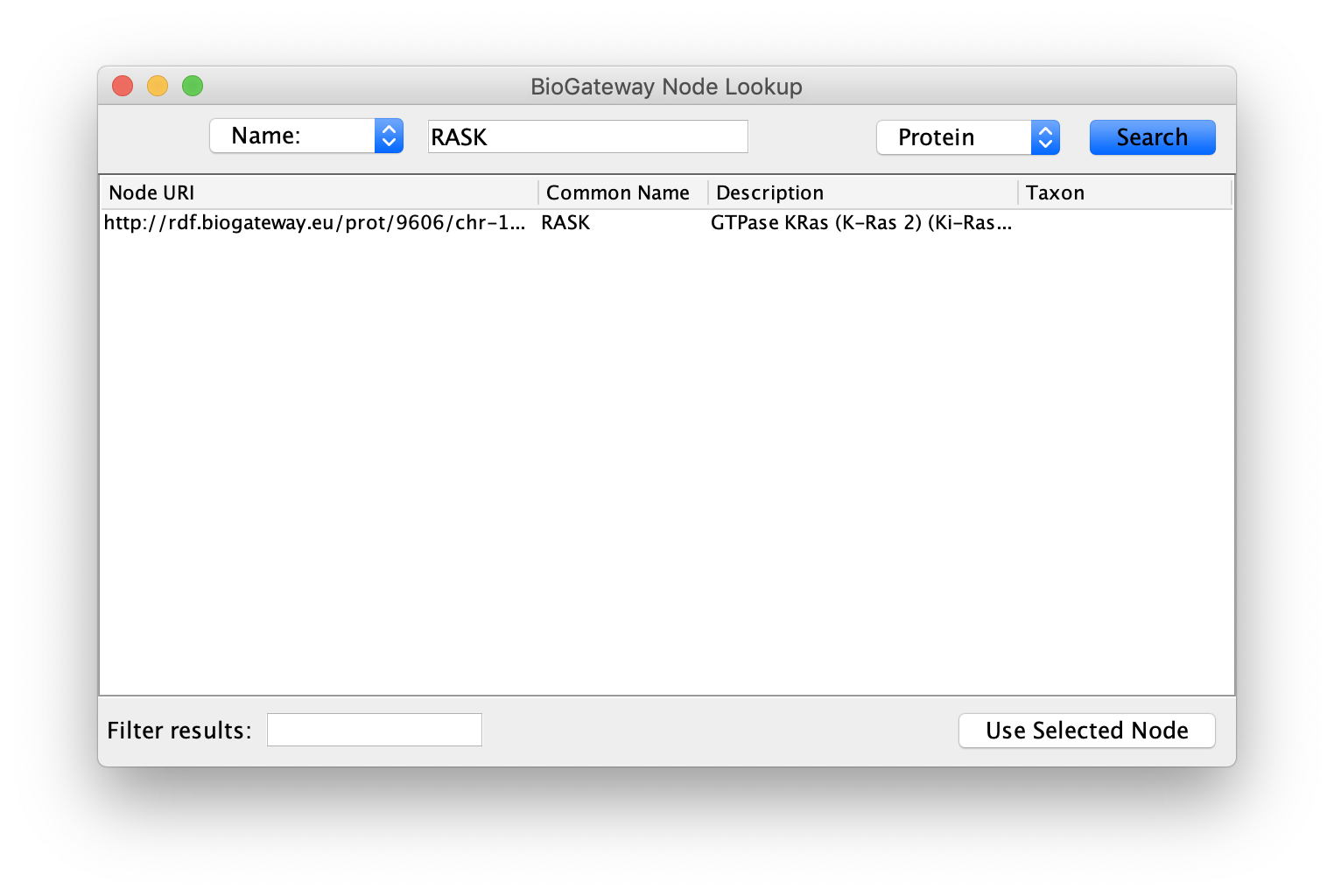
Starting from an empty network, the first step will be to right-click on the canvas and select BioGateway > Add a BioGateway node. Next, right-click on the canvas and select Biogateway > Add Biogateway node. The next step will be to enter the protein name RASK (all Capitals) in the query box (top of the BioGateway Node Lookup box, click Search, select the RASK protein and import by clicking Use Selected Node.
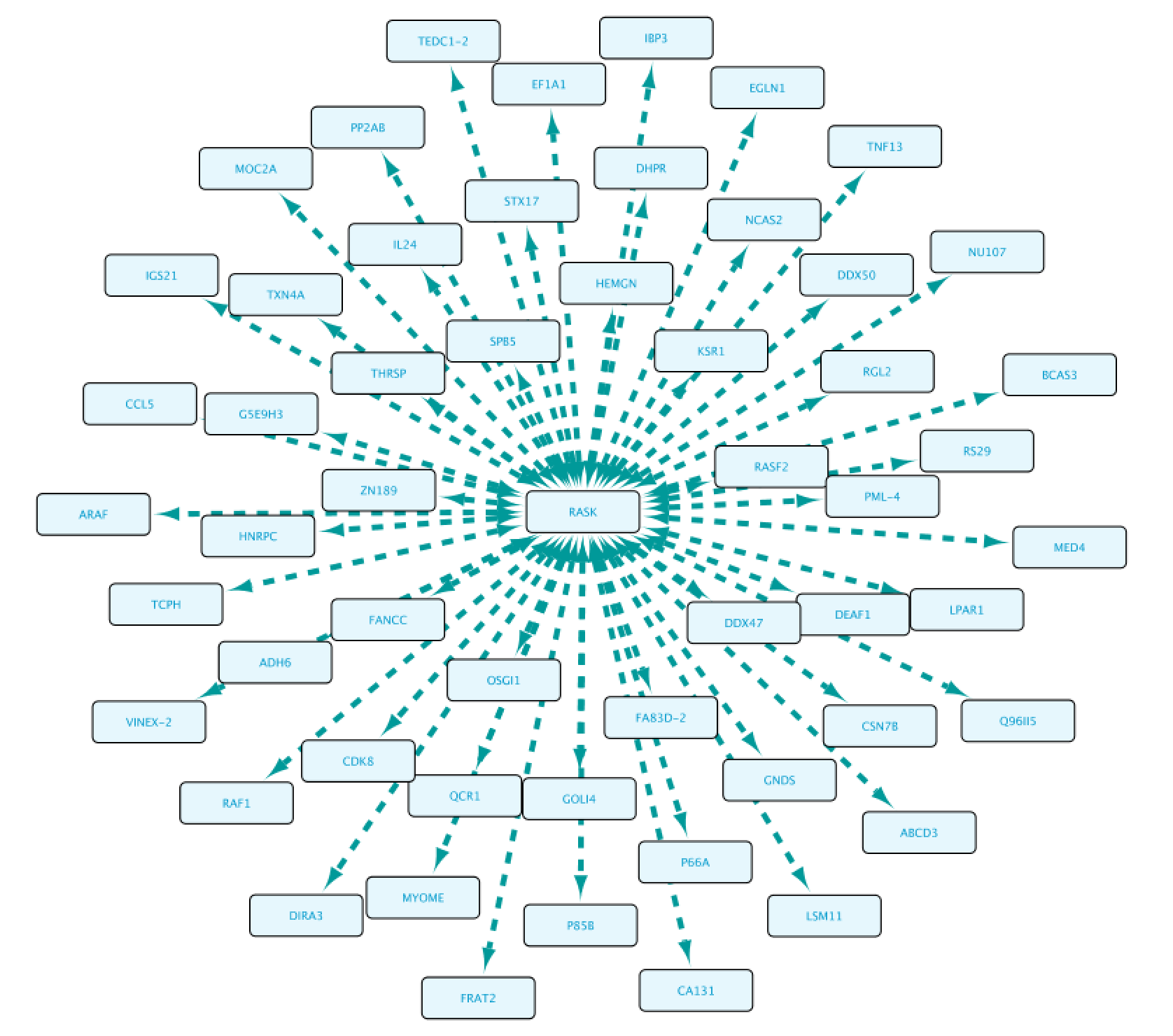
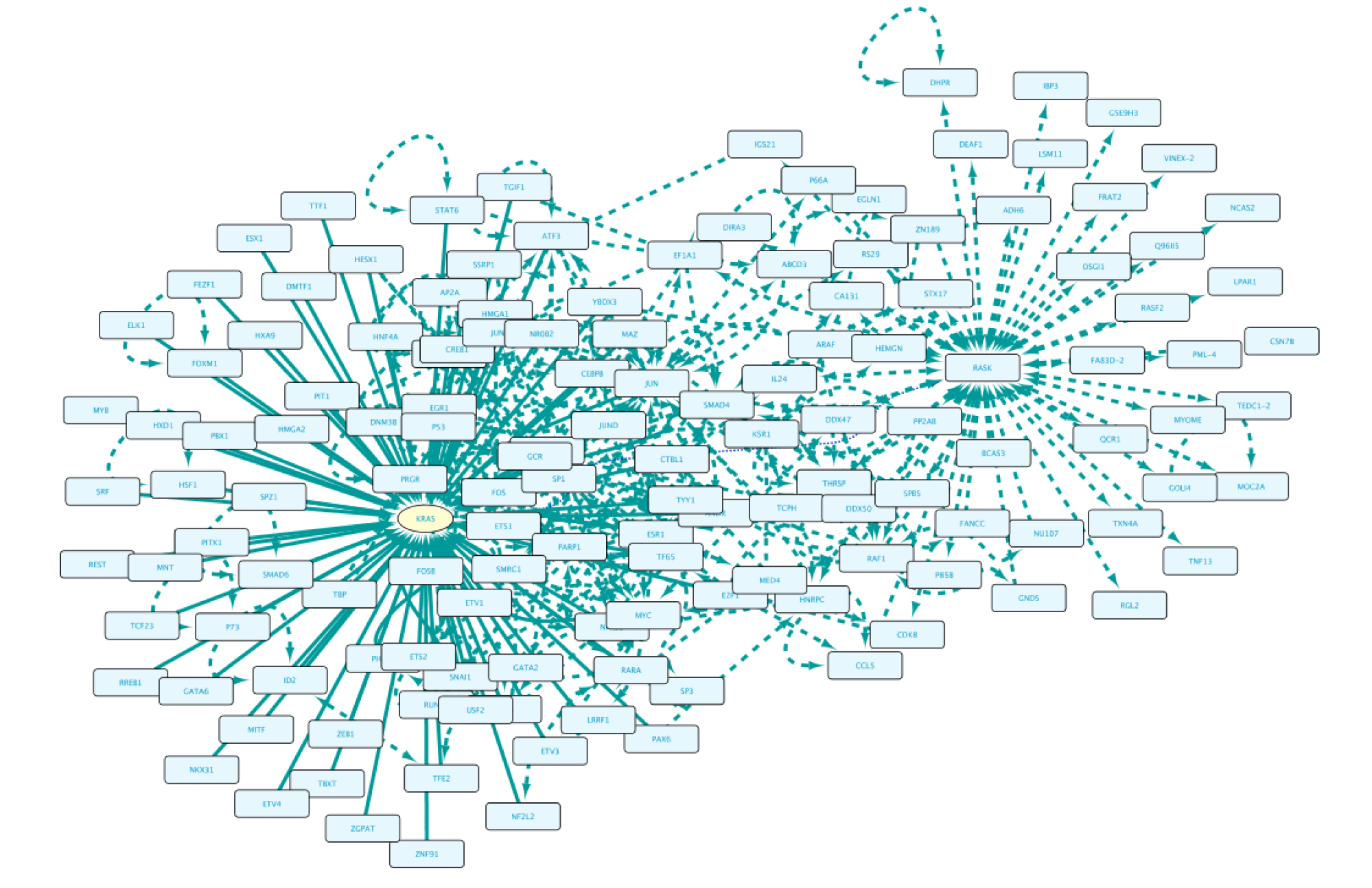
Once completed, right-click on the node in the Cytoscape network panel and expand, choosing from several options. In this step we select protein interaction partners from IntAct (right-click RASK, select ‘Biogateway’ > Fetch relations FROM node > IntAct: molecularly interacts with). Select all results and press Import Selected to display them to a network, adjust the display when necessary (e.g. Prefuse Force Directed Layout). The resulting network is shown below.
Now we will get the gene encoding for RASK and the TFs regulating it. To do so, right click on RASK and select Biogateway > Fetch relations TO node > UniProt Gene: encodes. Import the result, which then appears in the graph as a yellow oval indicating the gene KRAS. Now, right click on the KRAS gene and select Biogateway > Fetch relations TO node > Prot2Gene: involved in regulation of. Next, import all the results.
The last step will be to update the display (Prefuse Force Directed Layout) and add PPI interactions between all nodes in our network. This is done by selecting all nodes, right-clicking and selecting Biogateway > Fetch relations FROM selected > intact: molecularly interacts with. Finally, select all results, click Import relations between existing nodes.
The final network can be seen below.
Non-BioGateway method:
- Get interactions of RASK from Intact. Alternatively, get a previously built network containing RASK from repositories such as Reactome or others.
- Find all TFs in the network. This can be done by finding GO annotations that relate proteins to a TF function.
- Get the TGs of the TFs: use repositories such as TRRUST or TFactS to get the TGs from the TFs in our network.
- Find PPIs between the TFs found in step 2 and the proteins in our network using IntAct.
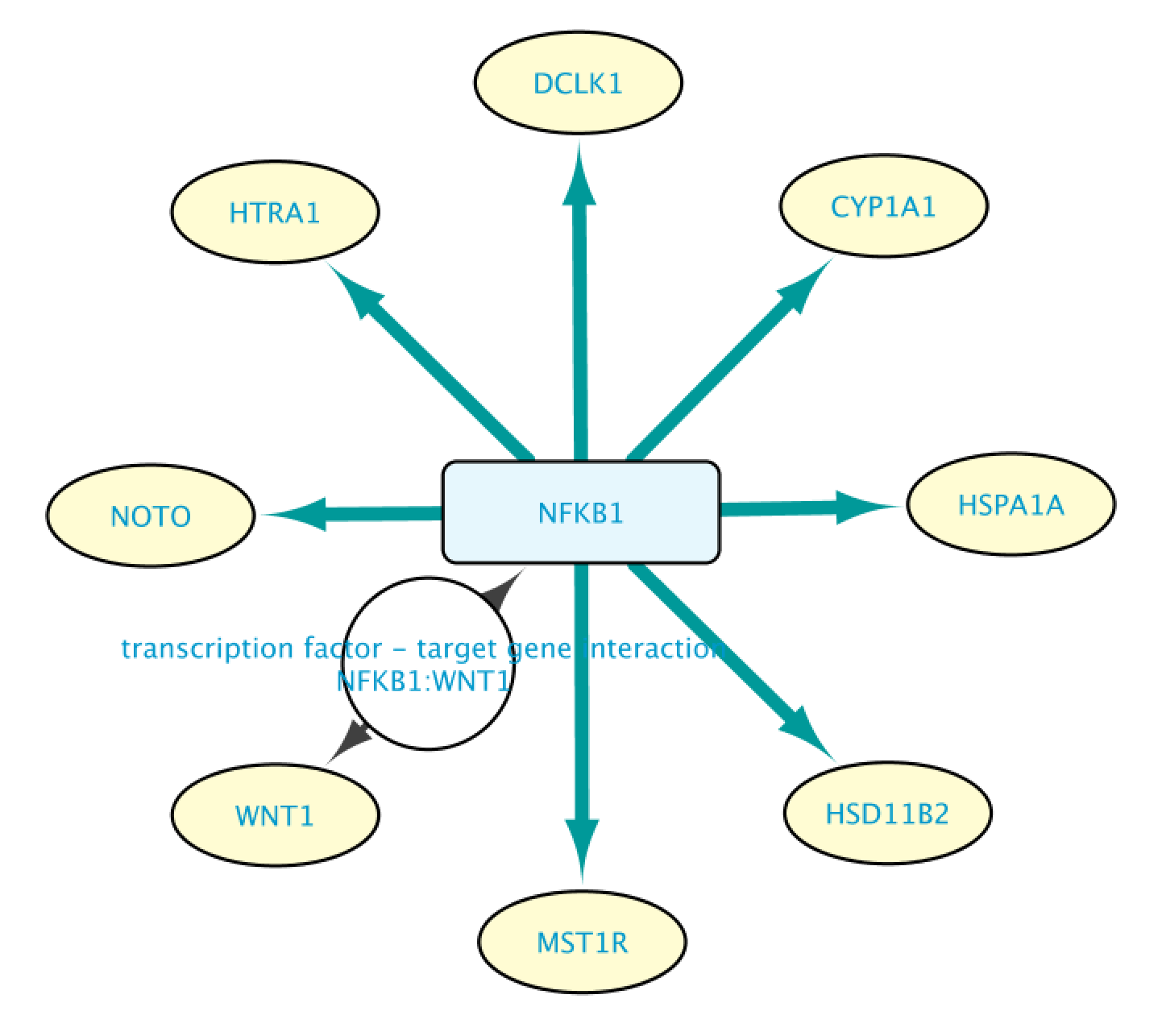
Use Case 2 - Building a TF-TG network starting from a TF of interest What genes are regulated by NFKB1? What is the literature supporting the results?
Create Query in Case2Query file (note that in the example we will only use a subset of the results). This query consists of one single line, where we ask for all the genes (Set A) whose transcription is regulated by NFKB1. Once we have defined the query, we click on the Run Query button.
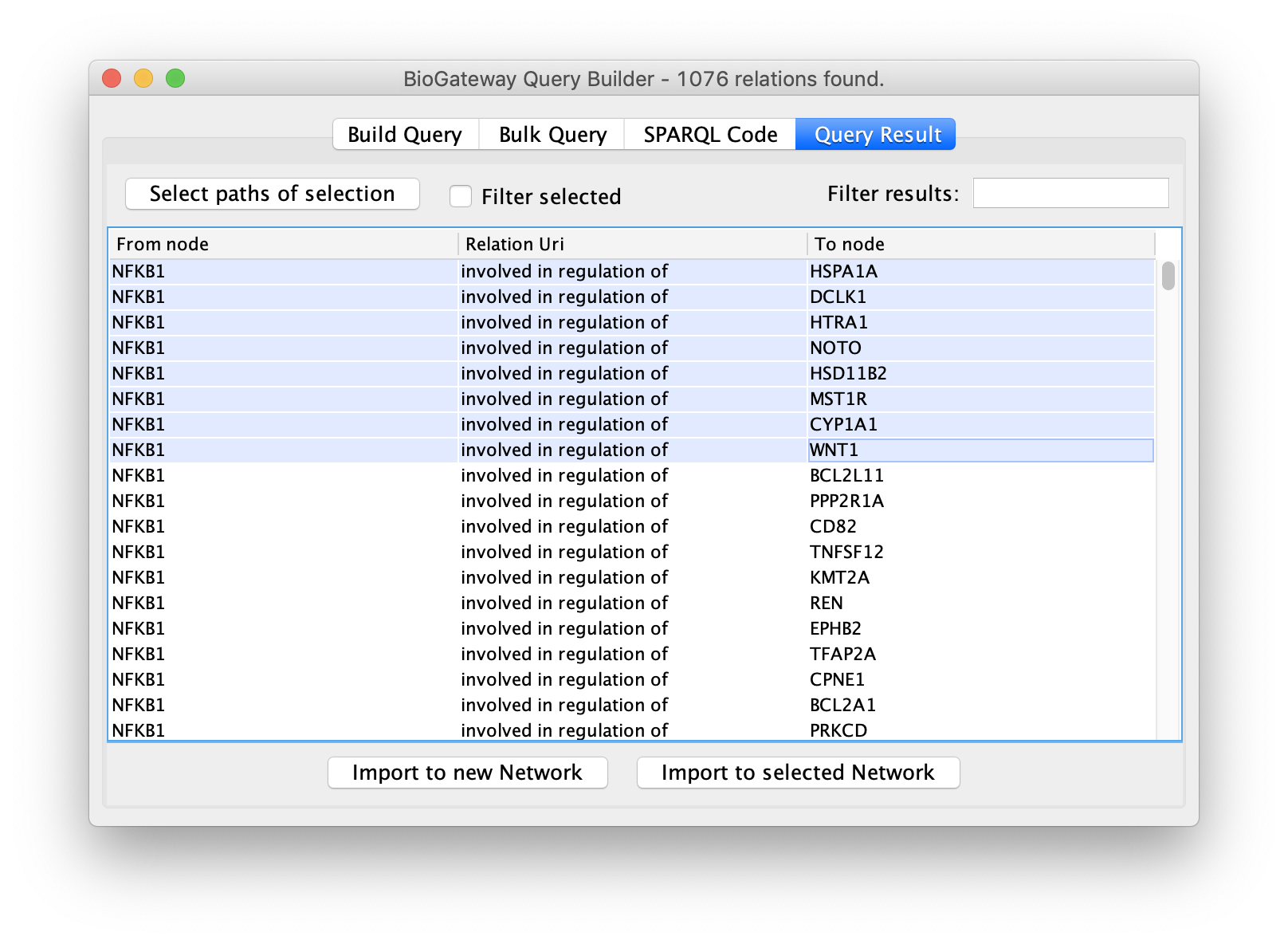
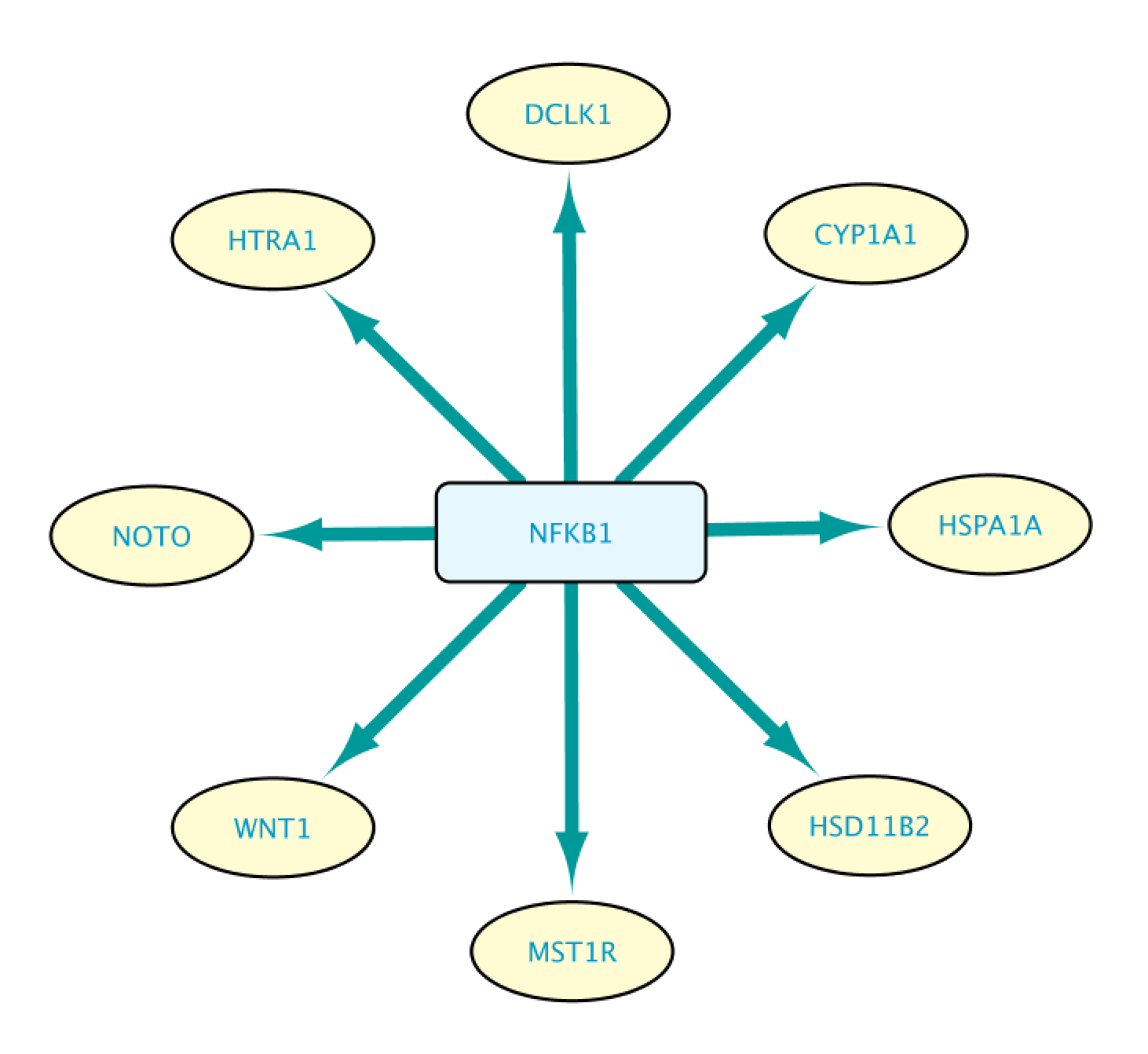
This will bring up the Query Result tab, where we select a subset of the target genes (HSPA1A, DCLK1, HTRA1, NOTO, HSD1182, MSTIR, CYP1A1 and WNT1). Next, we import the selected results by clicking on Import to selected Network, creating the network shown on the right.
We can expand an interaction by double clicking on it. This will let us see more information supporting that interaction statement. The figure below shows the network after expanding the interaction between NFKB1 and WNT1.
Right-clicking on the new node and following Biogateway > Open resource URI opens a tab in the web browser showing more data about that specific interaction.
The top panel shows the identity of the TF and TG, normalised to standard Uniprot and HGNC names, with a relationship provided by SIO (Semanticscience integrated Ontology). The bottom panel shows the abstract, with highlighted in purple the sentence that was found by text mining (EXTRI resource). The top-left of the bottom panel shows links to PubMed and the Europe PMC annotation platform.

Non-BioGateway method:
- Get TGs of a TF from a repository such as TRRUST.
- Check the publications supporting a TFTG statement.
Further checking of the supporting evidence:
Whereas the Landing page provides an initial documentation of the evidence that supports why the TF-TG relationship was included in the BioGateway Prot2Gene graph, a user may link out to the PubMed abstract (PMID ID) or to Europe PMC, which will (soon) provide access to the annotated abstract through their SciLite curation platform (see figure below).
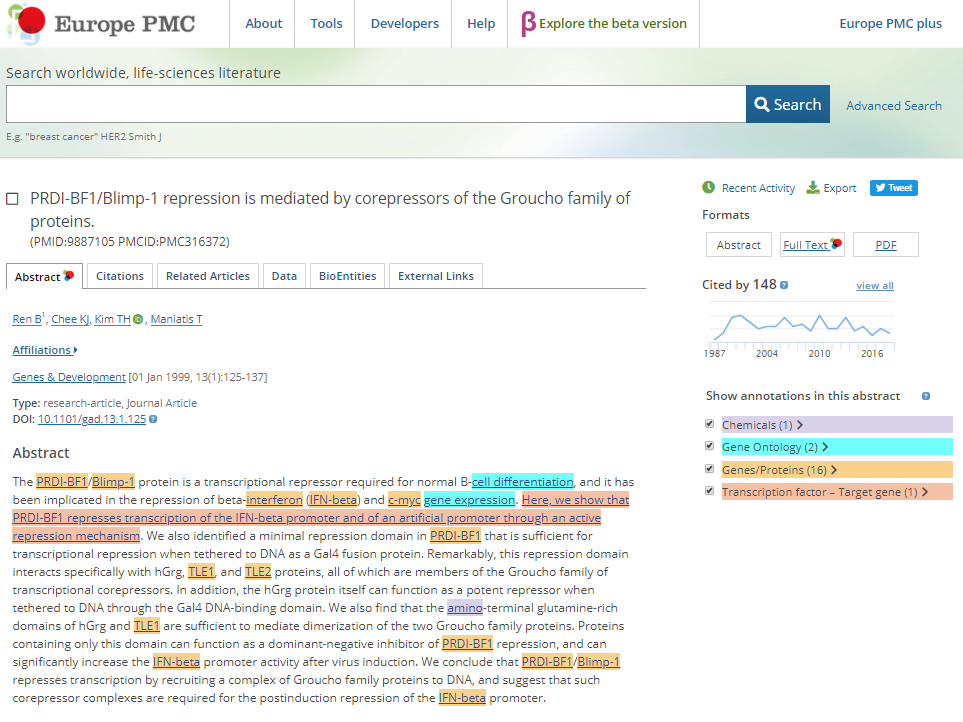
Europe PMC is developing the annotation platform SciLite. In the arbitrary example figure above, SciLite annotations are shown as highlighted pre-markings of chemicals, Gene Ontology terms, Genes and proteins, and a sentence that is part of the EXTRI resource (beta-version). The EXTRI effort will soon produce a new, high quality resource that we will upload in BioGateway. The sentence information will also be uploaded to the SciLite platform, where a user may indicate whether the sentence represents a bone-fide TF-TG relationship. The results of such a ‘community-curation’ effort will be incorporated via API back into BioGateway, where the confidence level (0, medium or high) will be updated.
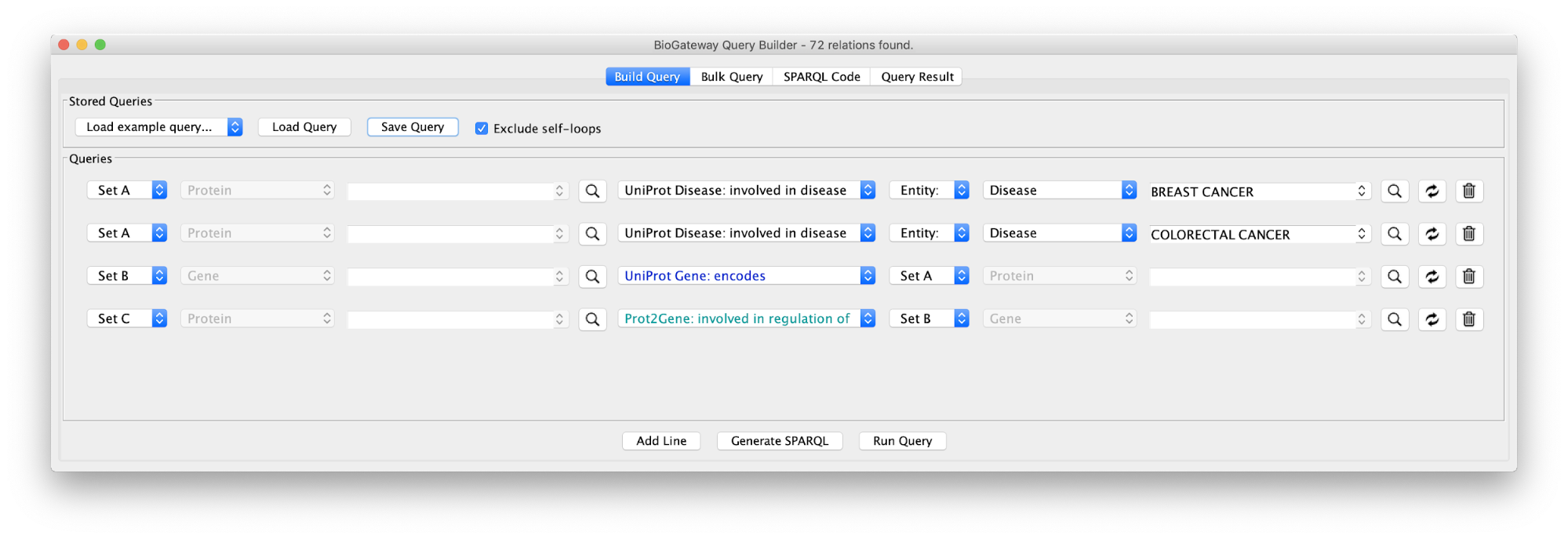
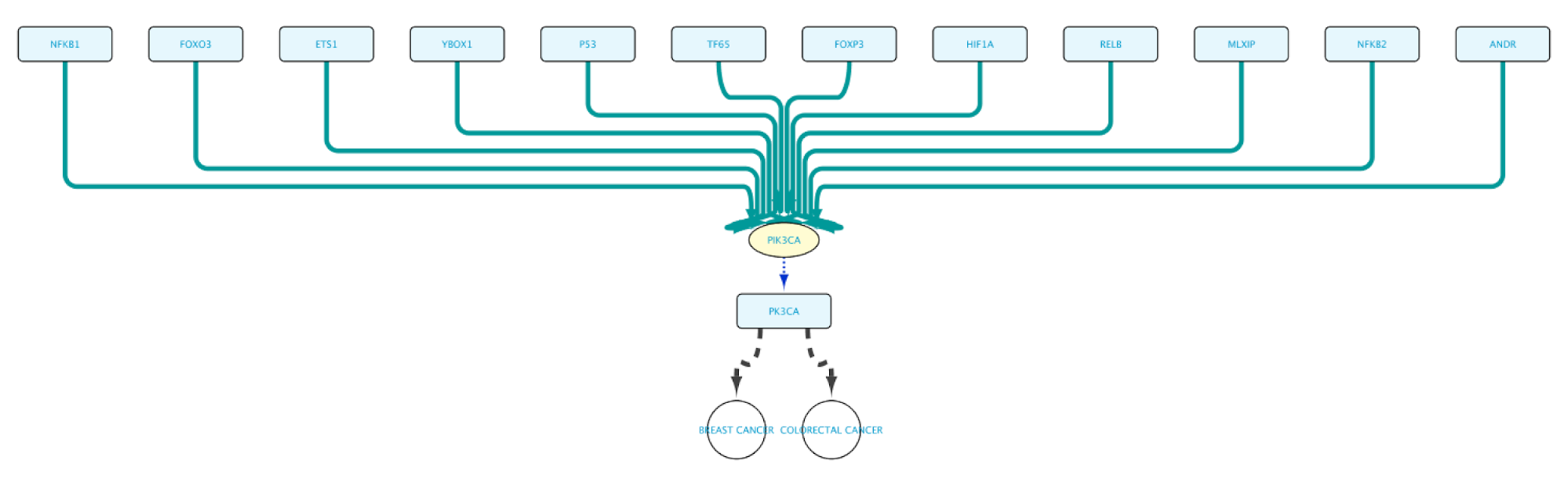
Use case 3 - Establishing Molecular Relationships between Diseases What proteins are involved in both Breast cancer and Colorectal cancer? What TFs regulate the transcription of the genes of these proteins?
Create the query below or import from file Case3Query, which will get all proteins known to be involved in both Breast cancer and Colorectal cancer, the genes encoding for those proteins and the Transcription Factors regulating these genes. The resulting network can be seen below.
Non-BioGateway method:
- Get proteins annotated to be involved in the desired diseases in a protein to disease repository.
- Find common proteins in both networks.
- Get the genes encoding for the found proteins in repositories like Entrez Gene.
- Get the TFs regulating these genes in repositories such as TRRUST or TFactS.
Use case 4 - Find the Proteins connecting two GO Terms Can we find the proteins that form a bridge between two distinct biological processes (cell adhesion to extracellular matrix and G1/S transition)?
The first step will be to create the query below, or load the query in file Case4Query.
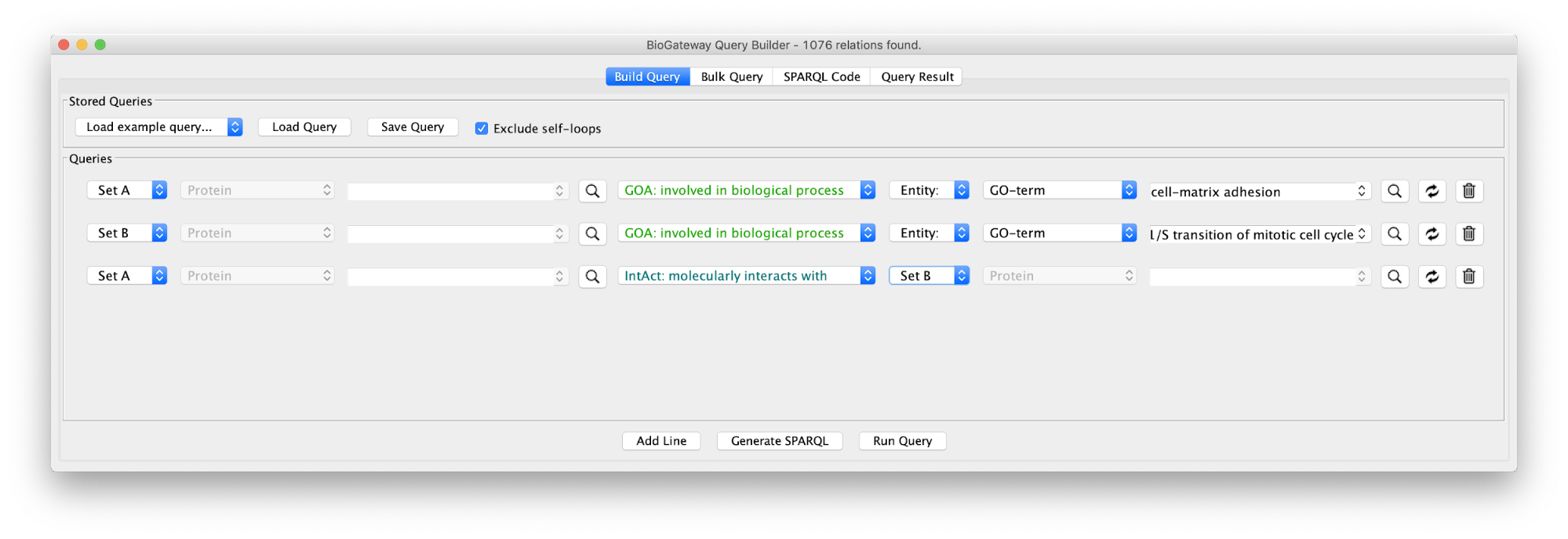
Running the query and importing all results will produce the following network:
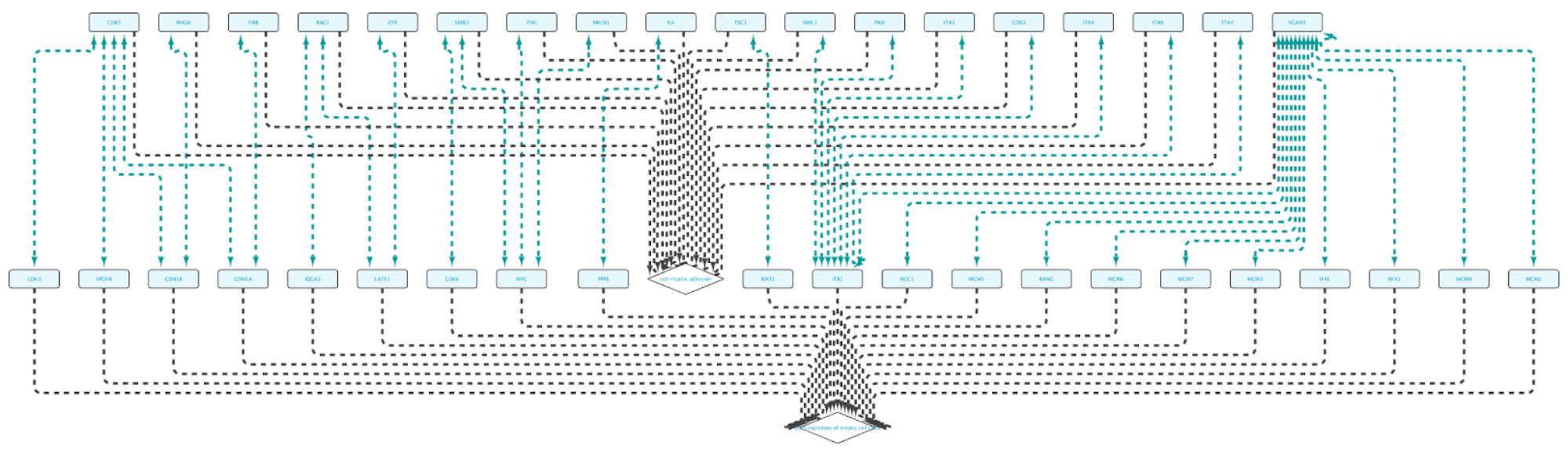
Non-BioGateway method:
- Find proteins annotated to the two GO terms separately.
- Look for common proteins to get the connections.
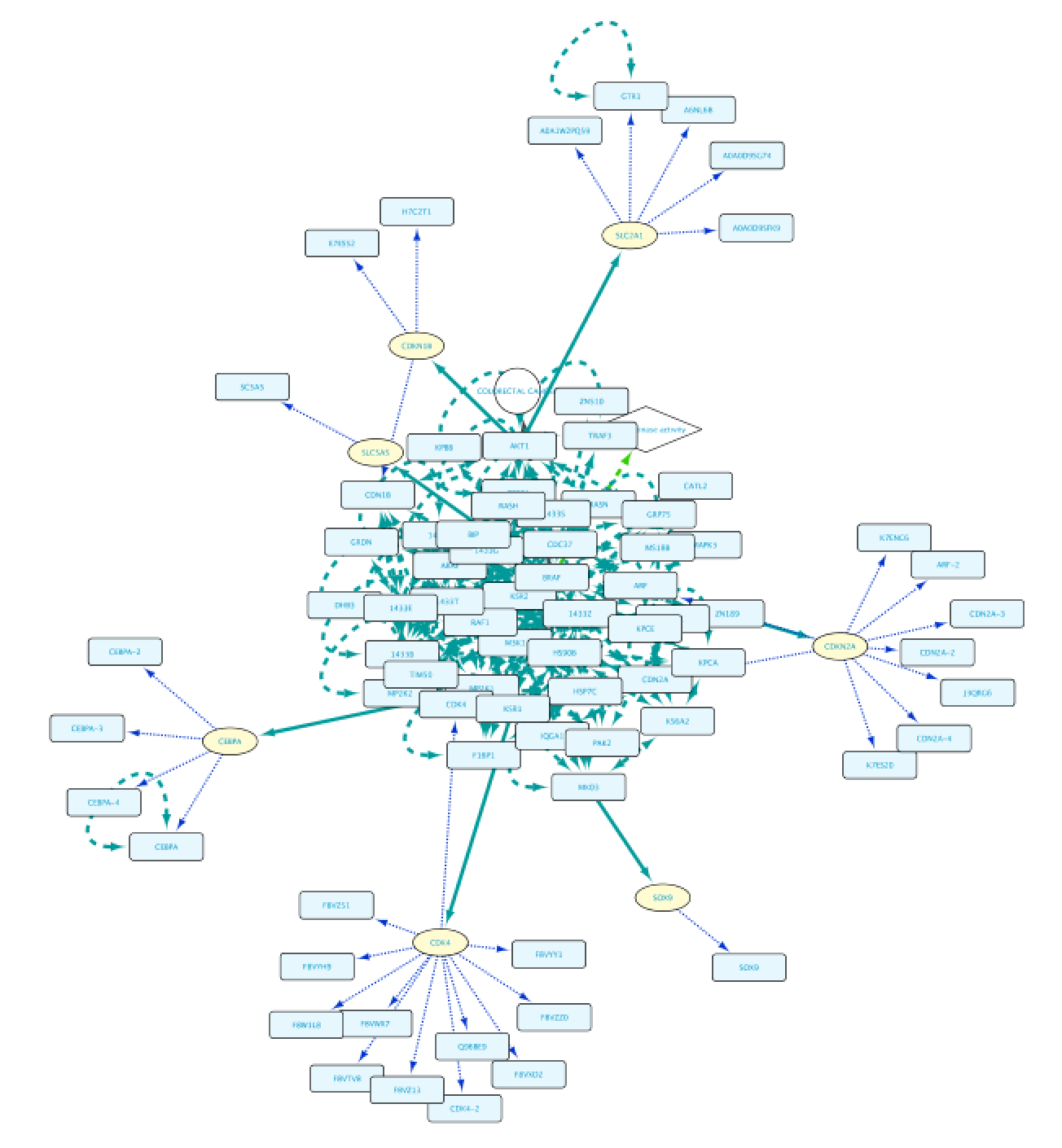
Use case 5 - Find proteins with protein kinase activity involved in a disease and the context around them What proteins with a kinase function are involved in colorectal cancer? What is the biological context around these proteins?
Build the query below or load the Case5Query from file, run the query and import all results.
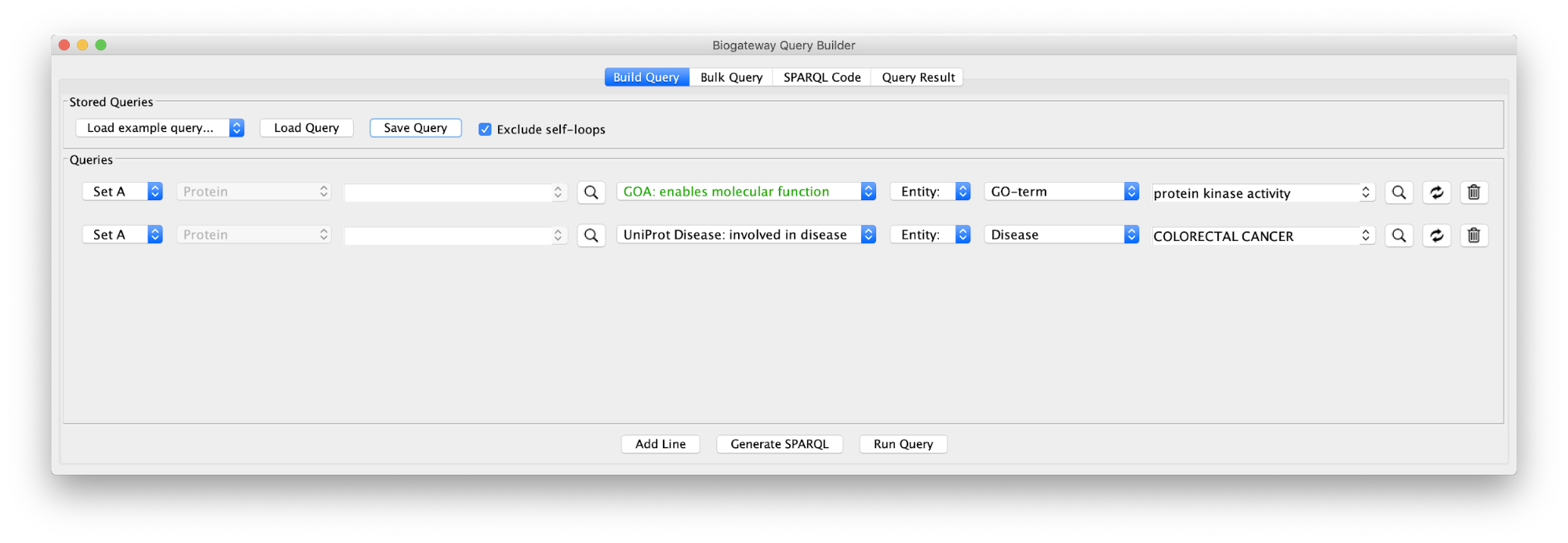
The result of this query can be seen in the image below: the protein BRAF, which has a protein kinase function and has an involvement in colorectal cancer. Now we will extend this small network by following a series of steps. The first one will be to get PPIs involving BRAF. Select this node and right-click Biogateway > Fetch relations FROM selected > IntAct: molecularly interacts with. Import all results. Next we will get the Target Genes of the Transcription Factors in the network. Select all nodes and Biogateway > Fetch relations FROM selected > Prot2Gene: involved in regulation of. Import all results.
The next step will be to get the proteins encoded by the regulated Target Genes and look for interactions with the existing proteins in the network. To do so, select all nodes and Biogateway > Fetch relations FROM selected > UniProt Gene: encodes. Import all results. Next, select all nodes again and Biogateway > Fetch relations FROM selected > IntAct: molecularly interacts with. Import relations between existing nodes.
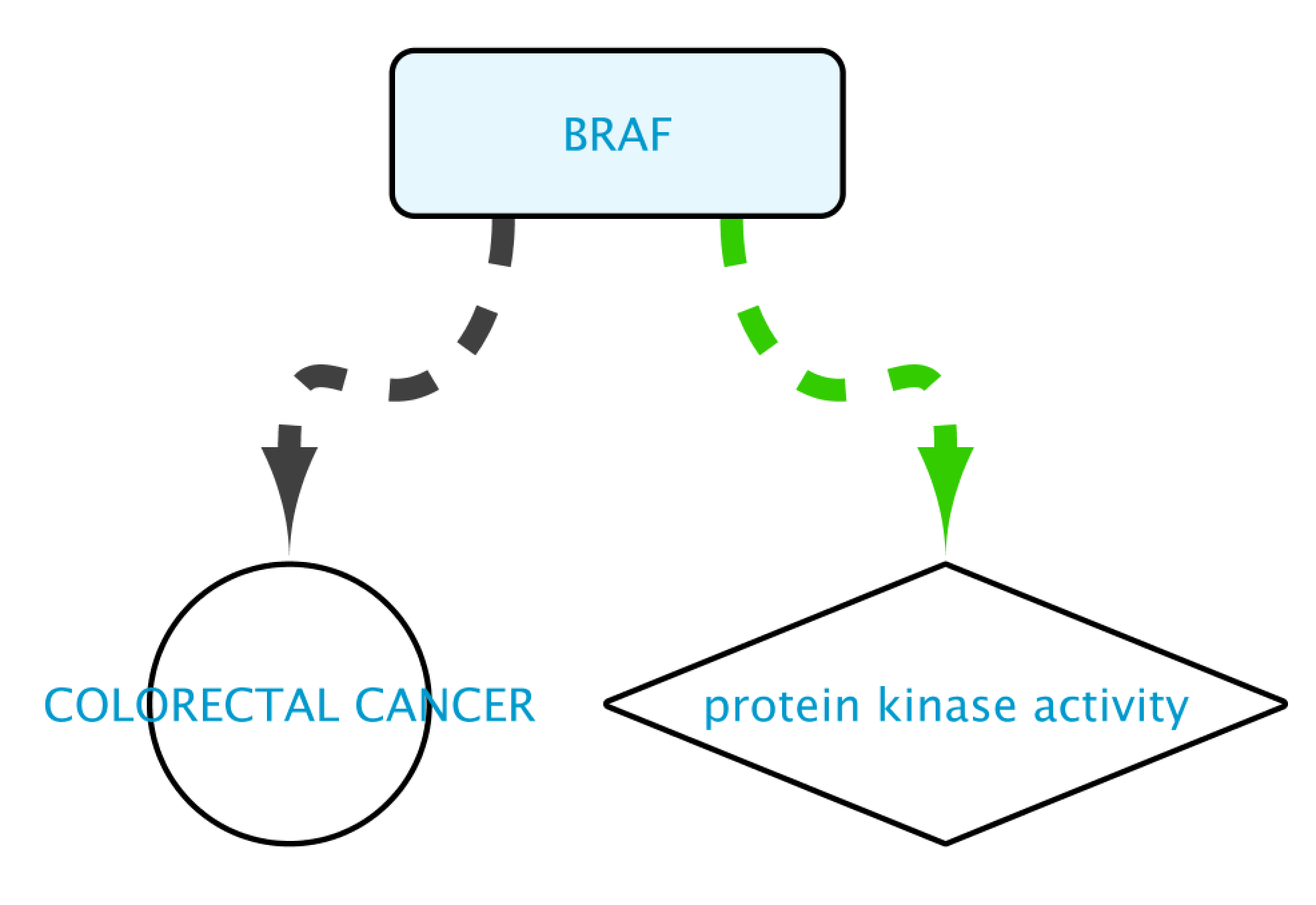
Non-BioGateway method:
- Find proteins annotated to the GOA protein kinase activity.
- Get proteins involved in diabetes-mellitus, non-insulin-dependent.
- Find common proteins in both networks.
- Find TGs of IRF7 in repositories like TRRUST, TFactS or others.
- Get proteins encoded by these TGs in Entrez Gene.
- Look for all PPIs involving the nodes in the network and find the ones involving the proteins in the built network.
Use case 5 (Extension) - Generalise a network from BioGateway for further analysis with Cytoscape apps What potential protein complexes can I find using MCODE on a network created by Biogateway?
We will extend the network from the use case 5. Thus, the first step will be to load, run and import all the results from the Case6Query. The next step will be to find all the genes regulated by the Transcription Factors in our network. Select all the nodes, right click and Biogateway > Fetch relations FROM selected > Prot2Gene: involved in regulation of. Import all relations.
Next, we will find the genes encoded by the genes in the network. Select all the nodes, right click and Biogateway > Fetch relations FROM selected > UniProt Gene: encodes. Import all relations. The following will be to find protein interactions between the proteins in our network. Select all the nodes, right click and Biogateway > Fetch relations FROM selected > IntAct: molecularly interacts with. Import connections between existing nodes.
Finally we will use the MCODE cytoscape plugin, which uses network analysis to look for potential complexes in a network. We will leave the default settings except that we will enable the option Include Loops. Next we will click on Analyze Current Network and create a sub-network with the first result.
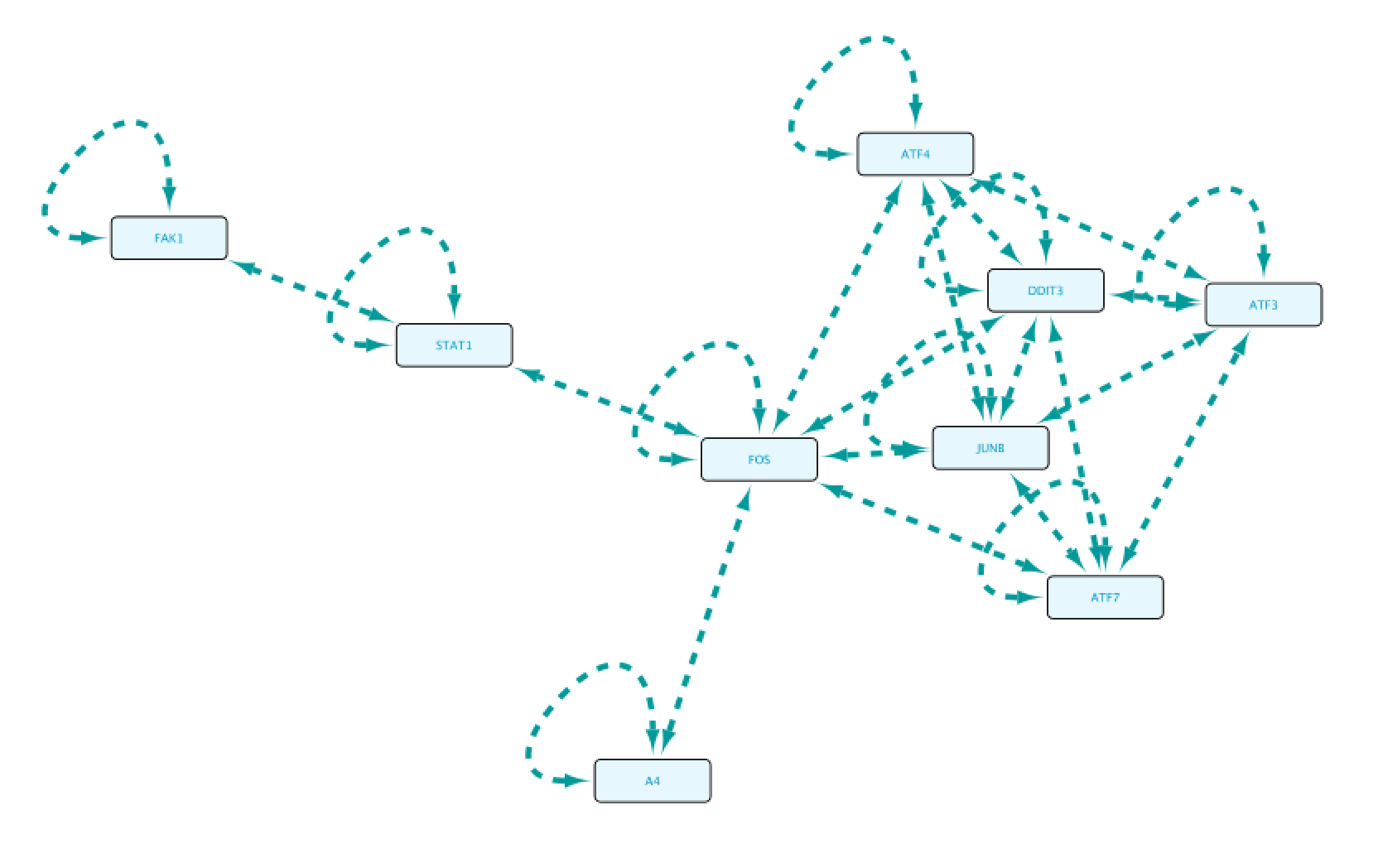
Use case 6 - Connect Protein A to Protein B (with GO restriction C) What PPIs can we find between proteins annotated to be involved in the G-protein coupled receptor signaling pathway?
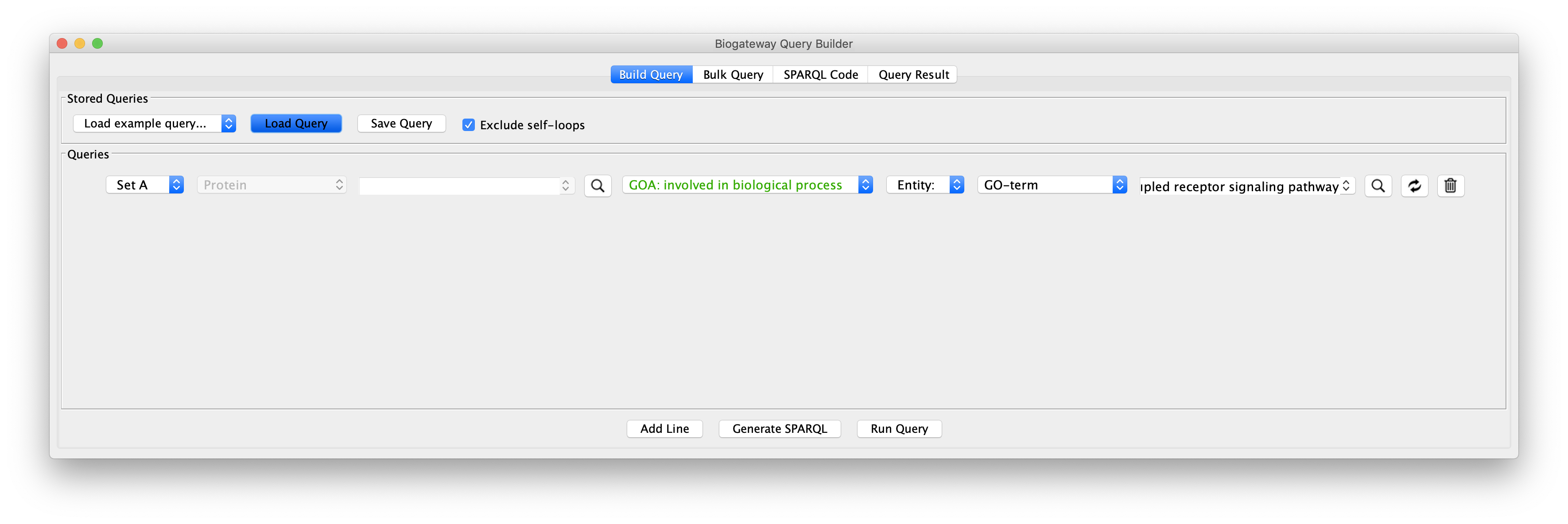
Create the query as stored in file Case6Query, which finds all proteins involved in a specific GO term. The example shows the GO term “G-protein coupled receptor signaling pathway”.
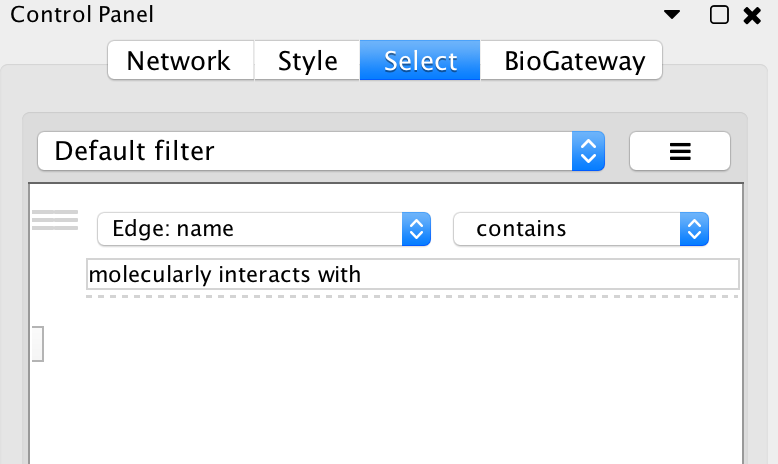
Now, we will look for PPIs between the nodes in the network: select all nodes and Biogateway > Fetch relations FROM selected > IntAct: molecularly interacts with. Import relations between existing nodes. Finally, we will create a subnetwork containing only the nodes connected by PPIs by using the Cytoscape filtering tool (Cytoscape Control panel > Select, see image below). This will create the subnetworks seen in the image on the right.
Non-BioGateway method:
- Find all proteins involved in the GO term of interest.
- Find connections between them on IntAct.
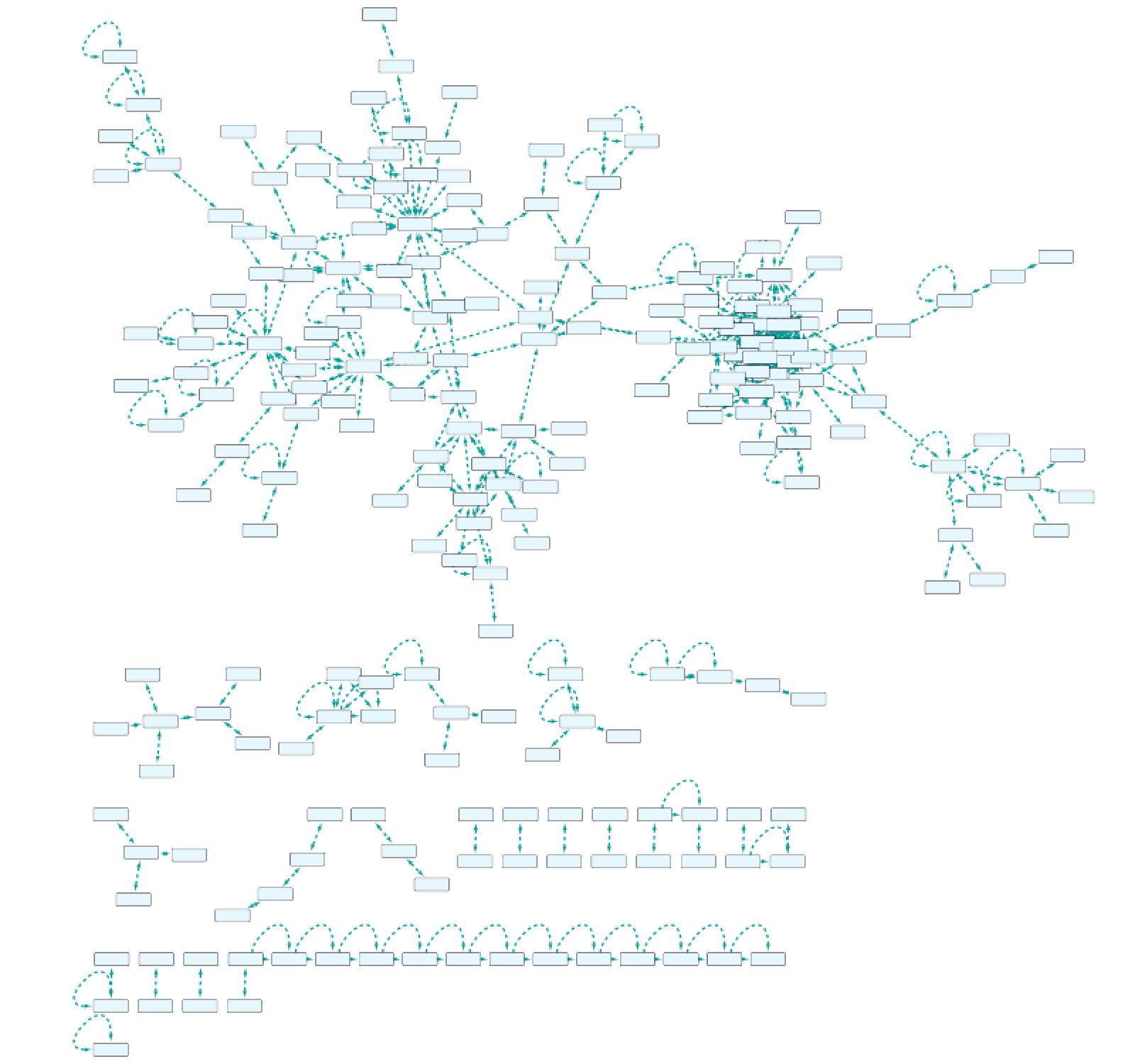
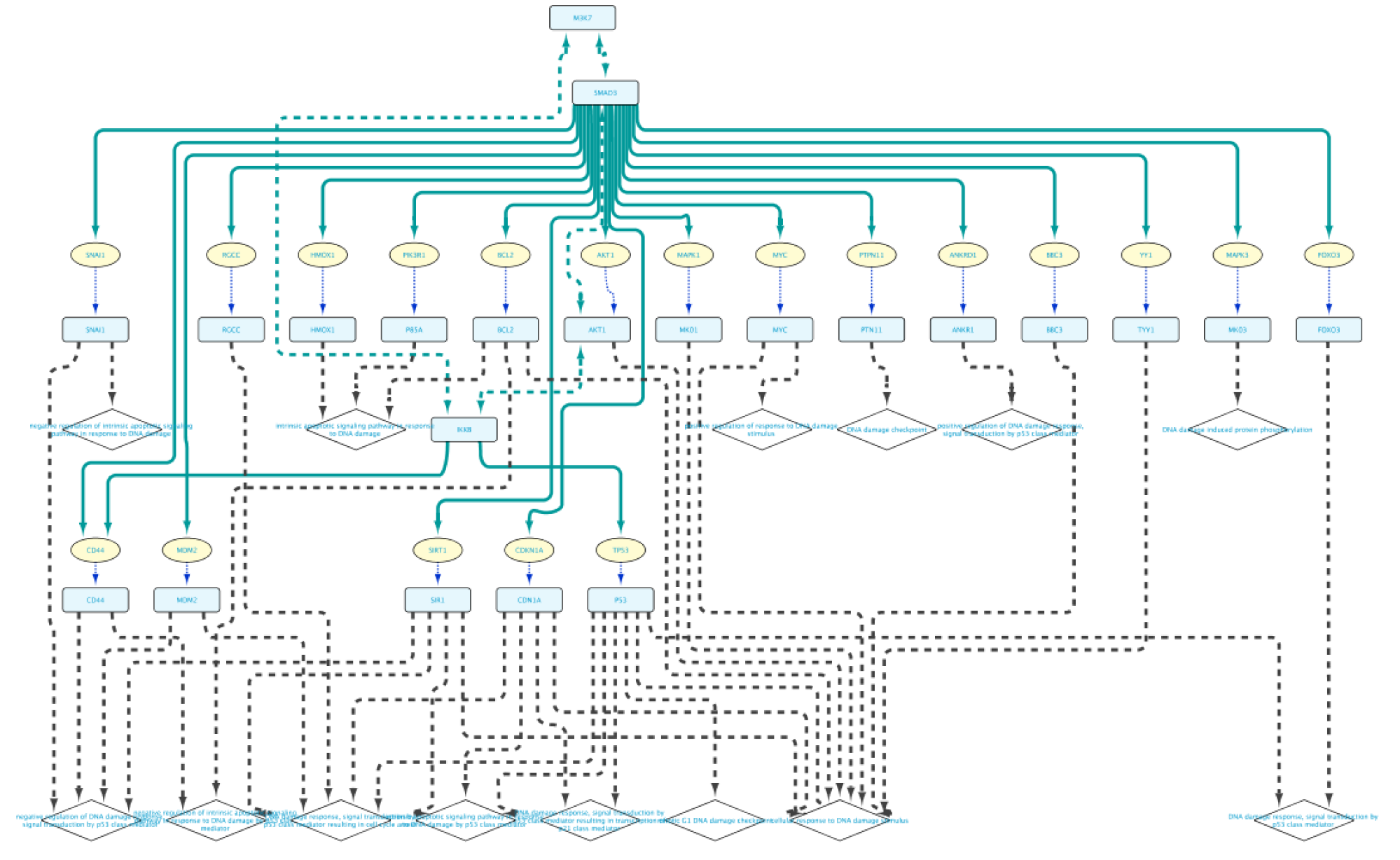
Use case 7 - Potential downstream effects after targeted inhibition of proteins What are the downstream effects of inhibiting MAP3K7 and AKT1? Can we find a connection between the two affected proteins?
Create the query below, as stored in the file Case7Query. You will notice that we have not specified a specific biological process term in line 5 of the query, which allows any annotation term in the results. Next, we can retrieve a subnetwork from the results by filtering for apoptotic process.
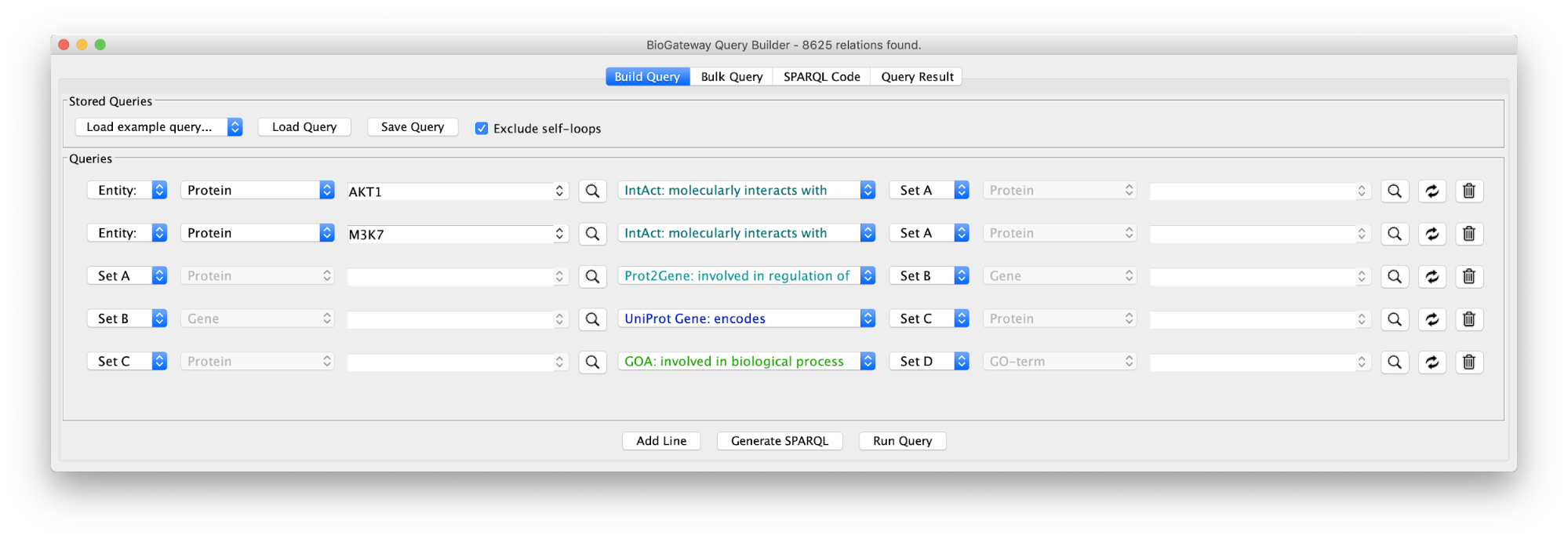
In the Query Results tab, we will use the Filter results text field, where writing DNA damage will select all GO biological process terms containing DNA damage.
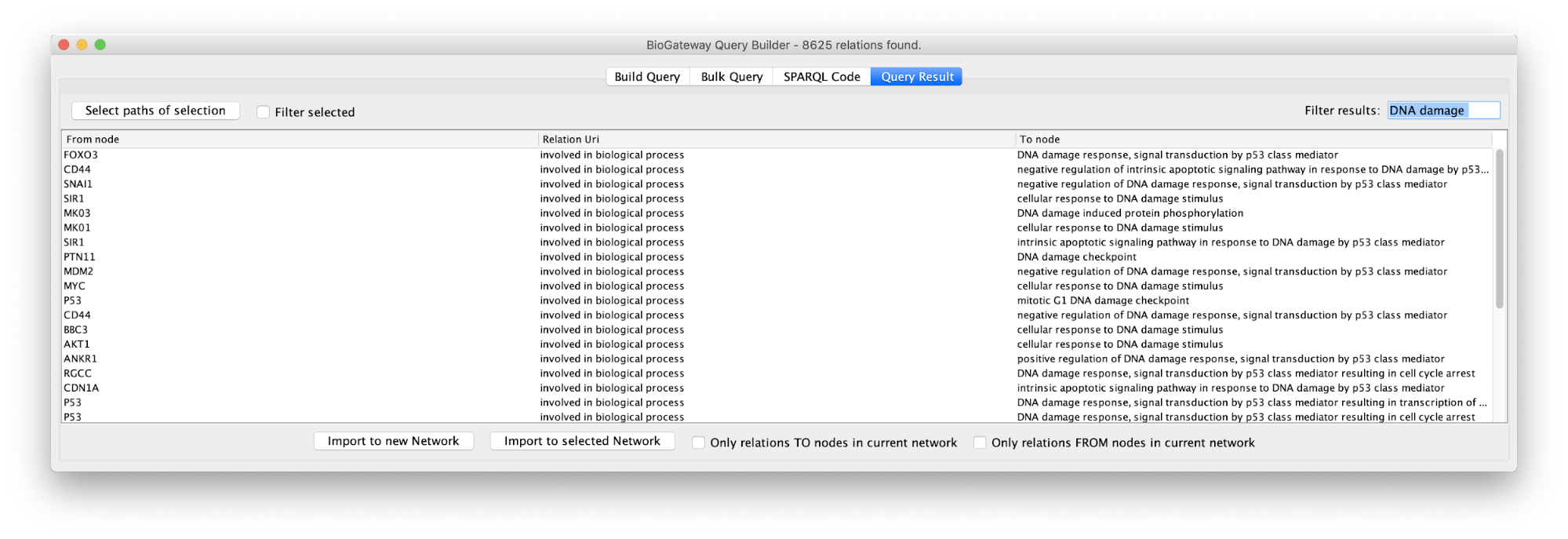
Now we will select all the relations in the results table and click on the Select paths of selection button on the top left corner. This will select only the relations creating the subnetwork we are interested in. Next, we click on Import to new Network. The resulting network can be seen on the right.
Non-BioGateway method:
- Check repositories of large-scale transcriptome data for transcriptome profiles measured after exposing cells to both drugs and perform functional enrichment analysis to reveal the affected biological process.
- Identify the targets in a protein-protein interaction (e.g., IntAct), identify which interacting partners are TFs (TFclass or GO [GO:0000981]), find the TGs (HTRIdb, TRRUST etc) and then, finally, check for the GO BP terms associated with the TGs (Uniprot, RefSeq, GO, GeneCards etc).