ontoPERL
An API for supporting the development and analysis of bio-ontologies
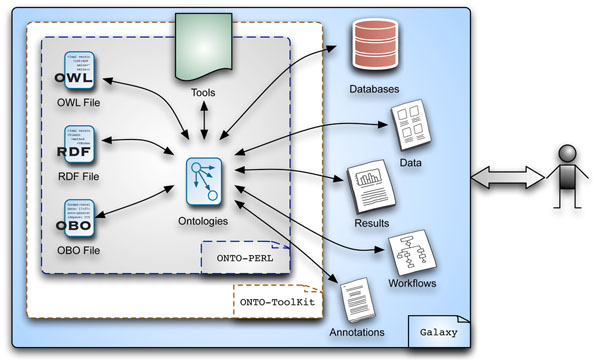
An Application Programming Interface (API) to programmatically deal with OBO/OWL based ontologies in many applications, including data integration, text mining, as well as semantic applications supporting translational research.
ONTO-ToolKit:
Enabling bio-ontology engineering via Galaxy
A functional integration of the Perl suite ONTO-PERL into the Galaxy platform. ONTO-ToolKit supports the analysis and handling of OBO-formatted ontologies via the Galaxy interface
Genes2GO
A web application for querying gene sets for specific GO terms
Genes2GO is an intuitive R-based web application that allows to retrieve custom sets of gene ontology annotations for any list of genes from organisms covered by the Ensembl database. The results are presented as a binary matrix file, indicating for each gene the presence or absence of specific annotations for a gene.
Link: http://norstore-trd-bio0.hpc.ntnu.no:8080/Genes2GO/
Cell Cycle Ontology
An application ontology for the representation and integrated analysis of the cell cycle process.
The Cell Cycle Ontology is an application ontology that automatically captures and integrates detailed knowledge on the cell cycle process.
Link to the CCO OWL file
Other link: http://bioportal.bioontology.org/ontologies/CCO
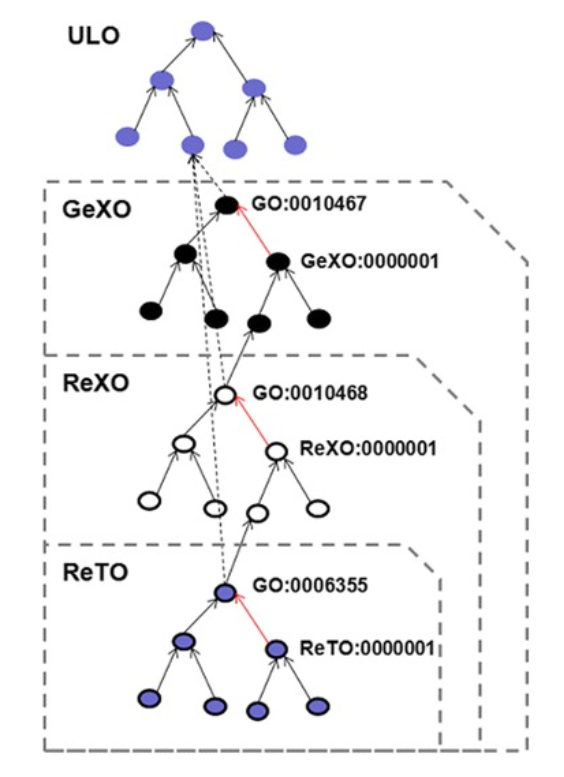
GeXO, ReXO and ReTO
Parts of the Gene expression Knowledge Base GeXKB
GeXO, ReXO and ReTO are nested, each ontologies connected to top nodes from the upper level ontology (ULO) the common root of the three ontologies. Black and red edges depict ‘is_a’ and ‘part_of’ relations, respectively. The three ontologies cover an increasingly wider domain. Each GO sub-domain term (e.g. GO:0010467; denoting ‘gene expression’) and its descendants are linked to the ULO as a subclass of ‘Biological Process’ represented by the ‘dotted edges’.
Paper:
Finding gene regulatory network candidates using the gene expression knowledge base
Links:
- http://bioportal.bioontology.org/ontologies/GEXO
- http://bioportal.bioontology.org/ontologies/REXO
- http://bioportal.bioontology.org/ontologies/RETO
LODQA Queries
The LODQA (Linked Open Data Question-Answering) project aims to automatically convert questions of biological interest, expressed in natural language, into SPARQL queries. LODQA provides a query interface to Linked Open Data resources with content that is indexed and ‘known’ to the LODQA query builder. The LODQA query interface is available here:
